Team:ETH Zurich/Project/Background
From 2008.igem.org
(→Top-down) |
|||
| (33 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
</center> | </center> | ||
</html> | </html> | ||
| - | + | {|style="background:#FFFFFF ; border:3.5px solid #60AFFE; padding: 1em; margin: auto ; width:98.5% " | |
| - | {|style="background:# | + | |
|- | |- | ||
| | | | ||
<!-- PUT THE PAGE CONTENT AFTER THIS LINE. THANKS :) --> | <!-- PUT THE PAGE CONTENT AFTER THIS LINE. THANKS :) --> | ||
| + | __TOC__ | ||
| + | ==Minimal genomes== | ||
| + | While the human genome contains around 30,000 genes (1), a worm needs | ||
| + | 19,099 genes (2) to survive and reproduce, while a fruit fly can live on just 13,600 genes (3). The smallest free-living eukaryote Ostreococcus tauri has a diminutive genome of just 8,166 genes (4), while baker’s yeast has a mere 6,563 genes. Organisms that do not indulge in the luxuries of multicellularity or a nucleus can live a happy life with even less genes. The standard laboratory bacterium Escherichia coli is happy with just 4,467 genes (5, 6) and bacteria that live parasitically inside other cells like Mycoplasma genitalium and Carsonella ruddii have record genomes of just 470 (7) and 182 (8) protein coding genes, respectively. | ||
| + | |||
| + | How many genes does a cell need in order to be able to survive and reproduce? Despite the wealth of genomic information that has been accumulated it is very difficult to answer this question in an absolute manner, since the number of genes required for survival is heavily depending on the conditions of growth. Single gene targeted deletion studies and the comparison between genomes shows that around 200 genes are absolutely essential (9), because they encode proteins that are needed to synthesize and handle DNA (such as DNA topoisomerases, DNA synthase), produce RNA (RNA polymerase) or translate the genetic information into proteins (ribosome, elongation factors, tRNAs). If the cell is provided with all nutrients required to build its DNA and proteins and has a source of energy, those approximately 200 genes are more or less all the cell needs for survival. Indeed, cells that live parasitically within eukaryotic or bacterial cells have some of the smallest genomes reported. | ||
| + | |||
| + | However, as soon as the cell lives in conditions that do not provide it with all the nutrients and energy it needs, it has to have additional genes that encode enzymes for biosynthesis of nutrients and energy conversion in order to survive and reproduce. If different sources of energy are available at different times, it will have to determine concentration of different compounds in the nutrient medium and react to changes in concentration (as E. coli does with the lac operon). If the environment grows more adverse, heat shock proteins, antifreeze proteins and other equipment might be necessary for survival. Therefore, the number of essential genes is directly linked to the growth conditions of the organism. | ||
| + | |||
| + | If a minimal genome for a certain condition is sought, there are in theory two ways to achieve it: bottom-up and top-down. In the bottom-up approach, the absolute minimal set of genes is used to begin with and additional genes are added on to enable the organism to survive under defined environmental conditions. Craig Venter’s recent complete synthesis of the Mycoplasma genitalium genome (10) and experiments towards transformation of whole genomes (11) take this direction. The top-down approach, on the other hand, would start with an established laboratory organism like E. coli and remove gene after gene either until the organism is not viable under the specified culture conditions (if a true minimal genome is sought, the last step before the organism becomes unviable would constitute the minimal genome for the specified culture conditions) or until the organism grows fastest (if elimination of evolutionary baggage and the result of a biotechnologically optimized genome is sought). | ||
| + | |||
| + | E. coli has been living in the intestine of animals for millennia and is perfectly adapted for this environment. Although it also grows extremely fast in shake flasks on standard culture media, the standard laboratory strains of E. coli are not adapted to the artificial environment in the sense that they still contain various biosynthetic pathways that are not needed for growth in standard glucose media and that therefore only add to the costs of protein synthesis without increasing the efficiency of the organism. Extensive research on the experimental evolution of E. coli by Richard E. Lenski have demonstrated that the growth rate of E. coli in shake flasks and glucose medium can already increase dramatically if cells are cultured for just 5,000 generations (13). During this process of adaptation, cells lose the ability to metabolize various energy sources that are not present in the medium and specialize on the sources that are provided. We could, therefore, postulate that it would be possible to create an E. coli with an optimized genome which has lost all biosynthetic pathways except for the ones necessary to metabolize the substances provided in the medium and synthesize the substances that are lacking. | ||
| + | |||
== Current approaches == | == Current approaches == | ||
| - | As usual when | + | As usual when tackling engineering projects, two main approaches can be followed: '''bottom-up''' and '''top-down'''. |
| + | |||
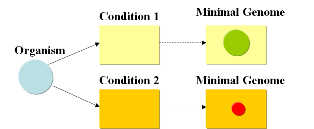
| + | We start with a given set of genes and the goal is to find the subset that contains the minimal amount of genes necessary to support life under given conditions. A specific medium containing metabolites needed for growth (such as LB) will require a different minimal set of genes than a minimal medium such as M9. This is due to the fact that a cell, living under conditions where metabolites are missing, will require additional genes that encode for the supplemental enzymes for biosynthesis and energy production in order to enable life (growth and reproduction). It will also need to have the possibility to adapt if different sources of energy are available at different times. | ||
| + | |||
| + | |||
| + | [[Image:conditions.png|center|frame|Fig. 1: Different conditions will result in a different set of minimal genes]] | ||
| + | |||
| + | |||
| + | ---- | ||
| + | '''A minimal genome is the minimal set of genes able to sustain life in a particular condition.''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | [[Image:geneset.png|center|200px]] | ||
| + | |||
| + | == Bottom-up == | ||
| + | |||
| + | In the first case we try to identify all the necessary functions for our system to work (i.e. to live). In this case, we identify pathways to produce all necessary metabolites the cell needs, such as lipids, aminoacids, etc. Here the chromosome of Mycoplasma genitalium with 580 kbp and 482 protein-coding genes has been selected as a starting point. Further analysis of the required functions has led to the selection of 382 genes in the chromosome. The following step is to synthesize the complete chromosome with the identified genes and transfer it into an "empty" cell (this approach is being followed e.g. by the JCVI). | ||
| + | |||
| + | [[Image:bottomup.png|center|200px]] | ||
| + | |||
| + | == Top-down == | ||
| + | |||
| + | The second approach starts from a working system (such as a well characterized strain like K12). By identifying non-essential parts of the metabolism and deleting them, we reduce the complexity of the cell. Many groups are working on this method, such as the Biofrontier Laboratories or Scarab Genomics. | ||
| + | |||
| + | |||
| + | [[Image:topdown.png|center|250px]] | ||
| + | |||
| + | |||
| + | A detailed descritption of our approach can be found in the [[Team:ETH Zurich/Project/Approach|approach]] section. | ||
| + | |||
| + | ==References== | ||
| + | |||
| + | |||
| + | (1) Lander, E. S. et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860-921. | ||
| + | |||
| + | (2) (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012-8. | ||
| + | |||
| + | (3) Adams, M. D. et al. (2000) The genome sequence of Drosophila melanogaster. Science 287, 2185-95. | ||
| + | (4) Derelle, E.et al. (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A 103, 11647-52. | ||
| + | |||
| + | (5) Blattner, F. R. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277, 1453-74. | ||
| + | |||
| + | (6) Riley, M. et al. (2006) Escherichia coli K-12: a cooperatively developed annotation snapshot--2005. Nucleic Acids Res 34, 1-9. | ||
| + | |||
| + | (7) Fraser, C. M. et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science 270, 397-403. | ||
| - | + | (8) Nakabachi, A. et al. (2006) The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314, 267. | |
| - | + | (9) Kobayashi, K. et al. (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A 100, 4678-83. | |
| + | (10) Gibson, D. G.et al. (2008) Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215-20. | ||
| + | (11) Lartigue, C.et al. (2007) Genome transplantation in bacteria: changing one species to another. | ||
| + | Science 317, 632-8. | ||
| + | (12) Posfai, G.et al. (2006) Emergent properties of reduced-genome Escherichia coli. Science 312, 1044-6. | ||
| + | (13) Cooper, V. S., and Lenski, R. E. (2000) The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407, 736-9. | ||
<!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | <!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | ||
|} | |} | ||
Latest revision as of 09:58, 30 October 2008
Minimal genomesWhile the human genome contains around 30,000 genes (1), a worm needs 19,099 genes (2) to survive and reproduce, while a fruit fly can live on just 13,600 genes (3). The smallest free-living eukaryote Ostreococcus tauri has a diminutive genome of just 8,166 genes (4), while baker’s yeast has a mere 6,563 genes. Organisms that do not indulge in the luxuries of multicellularity or a nucleus can live a happy life with even less genes. The standard laboratory bacterium Escherichia coli is happy with just 4,467 genes (5, 6) and bacteria that live parasitically inside other cells like Mycoplasma genitalium and Carsonella ruddii have record genomes of just 470 (7) and 182 (8) protein coding genes, respectively. How many genes does a cell need in order to be able to survive and reproduce? Despite the wealth of genomic information that has been accumulated it is very difficult to answer this question in an absolute manner, since the number of genes required for survival is heavily depending on the conditions of growth. Single gene targeted deletion studies and the comparison between genomes shows that around 200 genes are absolutely essential (9), because they encode proteins that are needed to synthesize and handle DNA (such as DNA topoisomerases, DNA synthase), produce RNA (RNA polymerase) or translate the genetic information into proteins (ribosome, elongation factors, tRNAs). If the cell is provided with all nutrients required to build its DNA and proteins and has a source of energy, those approximately 200 genes are more or less all the cell needs for survival. Indeed, cells that live parasitically within eukaryotic or bacterial cells have some of the smallest genomes reported. However, as soon as the cell lives in conditions that do not provide it with all the nutrients and energy it needs, it has to have additional genes that encode enzymes for biosynthesis of nutrients and energy conversion in order to survive and reproduce. If different sources of energy are available at different times, it will have to determine concentration of different compounds in the nutrient medium and react to changes in concentration (as E. coli does with the lac operon). If the environment grows more adverse, heat shock proteins, antifreeze proteins and other equipment might be necessary for survival. Therefore, the number of essential genes is directly linked to the growth conditions of the organism. If a minimal genome for a certain condition is sought, there are in theory two ways to achieve it: bottom-up and top-down. In the bottom-up approach, the absolute minimal set of genes is used to begin with and additional genes are added on to enable the organism to survive under defined environmental conditions. Craig Venter’s recent complete synthesis of the Mycoplasma genitalium genome (10) and experiments towards transformation of whole genomes (11) take this direction. The top-down approach, on the other hand, would start with an established laboratory organism like E. coli and remove gene after gene either until the organism is not viable under the specified culture conditions (if a true minimal genome is sought, the last step before the organism becomes unviable would constitute the minimal genome for the specified culture conditions) or until the organism grows fastest (if elimination of evolutionary baggage and the result of a biotechnologically optimized genome is sought). E. coli has been living in the intestine of animals for millennia and is perfectly adapted for this environment. Although it also grows extremely fast in shake flasks on standard culture media, the standard laboratory strains of E. coli are not adapted to the artificial environment in the sense that they still contain various biosynthetic pathways that are not needed for growth in standard glucose media and that therefore only add to the costs of protein synthesis without increasing the efficiency of the organism. Extensive research on the experimental evolution of E. coli by Richard E. Lenski have demonstrated that the growth rate of E. coli in shake flasks and glucose medium can already increase dramatically if cells are cultured for just 5,000 generations (13). During this process of adaptation, cells lose the ability to metabolize various energy sources that are not present in the medium and specialize on the sources that are provided. We could, therefore, postulate that it would be possible to create an E. coli with an optimized genome which has lost all biosynthetic pathways except for the ones necessary to metabolize the substances provided in the medium and synthesize the substances that are lacking. Current approachesAs usual when tackling engineering projects, two main approaches can be followed: bottom-up and top-down. We start with a given set of genes and the goal is to find the subset that contains the minimal amount of genes necessary to support life under given conditions. A specific medium containing metabolites needed for growth (such as LB) will require a different minimal set of genes than a minimal medium such as M9. This is due to the fact that a cell, living under conditions where metabolites are missing, will require additional genes that encode for the supplemental enzymes for biosynthesis and energy production in order to enable life (growth and reproduction). It will also need to have the possibility to adapt if different sources of energy are available at different times.
A minimal genome is the minimal set of genes able to sustain life in a particular condition.
Bottom-upIn the first case we try to identify all the necessary functions for our system to work (i.e. to live). In this case, we identify pathways to produce all necessary metabolites the cell needs, such as lipids, aminoacids, etc. Here the chromosome of Mycoplasma genitalium with 580 kbp and 482 protein-coding genes has been selected as a starting point. Further analysis of the required functions has led to the selection of 382 genes in the chromosome. The following step is to synthesize the complete chromosome with the identified genes and transfer it into an "empty" cell (this approach is being followed e.g. by the JCVI). Top-downThe second approach starts from a working system (such as a well characterized strain like K12). By identifying non-essential parts of the metabolism and deleting them, we reduce the complexity of the cell. Many groups are working on this method, such as the Biofrontier Laboratories or Scarab Genomics.
References(1) Lander, E. S. et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860-921. (2) (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012-8. (3) Adams, M. D. et al. (2000) The genome sequence of Drosophila melanogaster. Science 287, 2185-95. (4) Derelle, E.et al. (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A 103, 11647-52. (5) Blattner, F. R. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277, 1453-74. (6) Riley, M. et al. (2006) Escherichia coli K-12: a cooperatively developed annotation snapshot--2005. Nucleic Acids Res 34, 1-9. (7) Fraser, C. M. et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science 270, 397-403. (8) Nakabachi, A. et al. (2006) The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314, 267. (9) Kobayashi, K. et al. (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A 100, 4678-83. (10) Gibson, D. G.et al. (2008) Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215-20. (11) Lartigue, C.et al. (2007) Genome transplantation in bacteria: changing one species to another. Science 317, 632-8. (12) Posfai, G.et al. (2006) Emergent properties of reduced-genome Escherichia coli. Science 312, 1044-6. (13) Cooper, V. S., and Lenski, R. E. (2000) The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407, 736-9. |
 "
"