Team:NTU-Singapore/Parts/Characterization of pLsrA-YFP
From 2008.igem.org
(→Characterization of plsrA-YFP) |
Lalala8585 (Talk | contribs) |
||
| (4 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
| - | ='''Characterization of plsrA-YFP'''= | + | ='''Characterization of our new part: plsrA-YFP'''= |
| - | + | Link to the registry for Biobrick part:[http://partsregistry.org/wiki/index.php?title=Part:BBa_K117008 BBa_K117008] <br> | |
| - | ==Procedure== | + | [[Image: NTU@iGEM_PLsrA_YFP_pic.JPG|center|thumb|400px| pLsrA promoter with YFP reporting gene]]<br> |
| + | <br> | ||
| + | =='''Procedure'''== | ||
1) The successfully ligated plasmid with pLsrA-YFP gene was first transformed into chemically competent LuxS(-) cells. <br><br> | 1) The successfully ligated plasmid with pLsrA-YFP gene was first transformed into chemically competent LuxS(-) cells. <br><br> | ||
| - | 2) The next day, one colony of cell with pLsrA-YFP plasmid was inoculated in 5ml LBA for 16 hours at | + | 2) The next day, one colony of cell with pLsrA-YFP plasmid was inoculated in 5ml LBA for 16 hours at 37°C and shaked at 225 rpm.<br><br> |
| - | 3) Overnight cell culture was then centrifuged at 4000 rpm and | + | 3) Overnight cell culture was then centrifuged at 4000 rpm and 4°C for 10 minutes. <br><br> |
4) The supernatant was discarded and cell pellets were re-suspended in 5ml of Ampicilin-containing M9 medium. The amount of M9 medium was adjusted until cell suspension had an OD600 of 1. <br><br> | 4) The supernatant was discarded and cell pellets were re-suspended in 5ml of Ampicilin-containing M9 medium. The amount of M9 medium was adjusted until cell suspension had an OD600 of 1. <br><br> | ||
5) The cell suspensions were then pipetted into 96-well microplate wells. This was followed by the adding of 50µl AI-2-containing supernatant into corresponding wells. <br><br> | 5) The cell suspensions were then pipetted into 96-well microplate wells. This was followed by the adding of 50µl AI-2-containing supernatant into corresponding wells. <br><br> | ||
| - | + | 7) There were 2 different negative control samples being used for this study.<br> | |
| - | + | ||
* The First control sample contained cell suspension only. <br> | * The First control sample contained cell suspension only. <br> | ||
| - | |||
* And as the supernatants also contain large amount of LB, cell suspension with 50µl <br> | * And as the supernatants also contain large amount of LB, cell suspension with 50µl <br> | ||
LB added was also used as another control sample. <br> | LB added was also used as another control sample. <br> | ||
<br> | <br> | ||
8) YFP measurement was carried out by VICTOR 3 multilabel reader at excitation wavelength of 490 nm and emission wavelength of 535 nm. <br><br> | 8) YFP measurement was carried out by VICTOR 3 multilabel reader at excitation wavelength of 490 nm and emission wavelength of 535 nm. <br><br> | ||
| - | 9) Data were automatically gathered every 10 minutes and temperature was set at | + | 9) Data were automatically gathered every 10 minutes and temperature was set at 37°C. |
<br><br> | <br><br> | ||
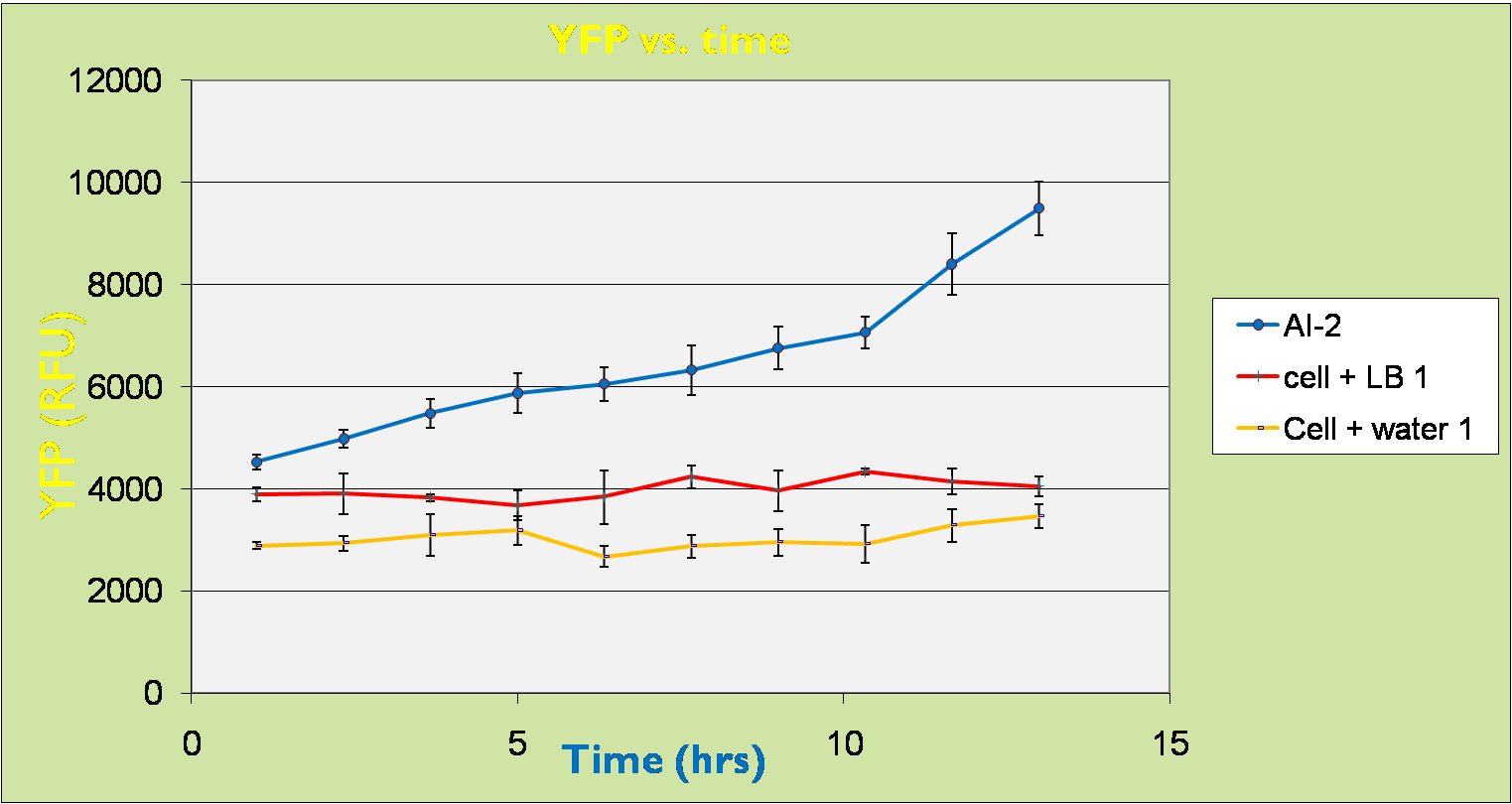
| - | [[Image: | + | [[Image:YFP time.png|thumb|center|800px|YFP vs Time Graph]]<br> |
| - | For | + | For the control samples with only cells, the RFU remained at a steady level with time. The same thing occurred for the cells with 50µl LB broth added, although RFU level was a bit higher. For the cells with 50µl supernatants containing AI-2, we can clearly observe an increasing trend in the RFU. |
This further proves that the LsrA promoter part BBa_K117002 in fact works correctly as it is activated in the presence of AI-2. Further characterization of this promoter will be implemented in future works. | This further proves that the LsrA promoter part BBa_K117002 in fact works correctly as it is activated in the presence of AI-2. Further characterization of this promoter will be implemented in future works. | ||
| - | From this result, it is highly expected that the detection system part BBa_K117010 also expresses lysis protein (celE7) upon induced by AI-2. | + | From this result, it is highly expected that the detection system part BBa_K117010 also expresses lysis protein (celE7) upon induced by AI-2. |
| + | <br><br> | ||
| + | <html> | ||
| + | <script language=Javascript1.2> | ||
| + | <!-- | ||
| + | |||
| + | var tags_before_clock = "<b>It is now " | ||
| + | var tags_middle_clock = "on" | ||
| + | var tags_after_clock = "</b>" | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.write('<layer id="clock"></layer><br>'); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | document.write('<span id="clock"></span>'); | ||
| + | } | ||
| + | |||
| + | DaysofWeek = new Array() | ||
| + | DaysofWeek[0]="Sunday" | ||
| + | DaysofWeek[1]="Monday" | ||
| + | DaysofWeek[2]="Tuesday" | ||
| + | DaysofWeek[3]="Wednesday" | ||
| + | DaysofWeek[4]="Thursday" | ||
| + | DaysofWeek[5]="Friday" | ||
| + | DaysofWeek[6]="Saturday" | ||
| + | |||
| + | Months = new Array() | ||
| + | Months[0]="January" | ||
| + | Months[1]="February" | ||
| + | Months[2]="March" | ||
| + | Months[3]="April" | ||
| + | Months[4]="May" | ||
| + | Months[5]="June" | ||
| + | Months[6]="July" | ||
| + | Months[7]="August" | ||
| + | Months[8]="September" | ||
| + | Months[9]="October" | ||
| + | Months[10]="November" | ||
| + | Months[11]="December" | ||
| + | |||
| + | function upclock(){ | ||
| + | var dte = new Date(); | ||
| + | var hrs = dte.getHours(); | ||
| + | var min = dte.getMinutes(); | ||
| + | var sec = dte.getSeconds(); | ||
| + | var day = DaysofWeek[dte.getDay()] | ||
| + | var date = dte.getDate() | ||
| + | var month = Months[dte.getMonth()] | ||
| + | var year = dte.getFullYear() | ||
| + | |||
| + | var col = ":"; | ||
| + | var spc = " "; | ||
| + | var com = ","; | ||
| + | var apm; | ||
| + | |||
| + | if (date == 1 || date == 21 || date == 31) | ||
| + | {ender = "<sup>st</sup>"} | ||
| + | else | ||
| + | if (date == 2 || date == 22) | ||
| + | {ender = "<sup>nd</sup>"} | ||
| + | else | ||
| + | if (date == 3 || date == 23) | ||
| + | {ender = "<sup>rd</sup>"} | ||
| + | |||
| + | else | ||
| + | {ender = "<sup>th</sup>"} | ||
| + | |||
| + | if (12 < hrs) { | ||
| + | apm="<font size='-1'>pm</font>"; | ||
| + | hrs-=12; | ||
| + | } | ||
| + | |||
| + | else { | ||
| + | apm="<font size='-1'>am</font>"; | ||
| + | } | ||
| + | |||
| + | if (hrs == 0) hrs=12; | ||
| + | if (hrs<=9) hrs="0"+hrs; | ||
| + | if (min<=9) min="0"+min; | ||
| + | if (sec<=9) sec="0"+sec; | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.clock.document.write(tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock); | ||
| + | document.clock.document.close(); | ||
| + | } | ||
| - | + | if (navigator.appVersion.indexOf("MSIE") != -1){ | |
| + | clock.innerHTML = tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock; | ||
| + | } | ||
| + | } | ||
| - | + | setInterval("upclock()",1000); | |
| - | + | //--> | |
| + | </script> | ||
| + | </html> | ||
Latest revision as of 05:50, 27 October 2008
|
Characterization of our new part: plsrA-YFP
Link to the registry for Biobrick part:[http://partsregistry.org/wiki/index.php?title=Part:BBa_K117008 BBa_K117008]
Procedure
1) The successfully ligated plasmid with pLsrA-YFP gene was first transformed into chemically competent LuxS(-) cells.
2) The next day, one colony of cell with pLsrA-YFP plasmid was inoculated in 5ml LBA for 16 hours at 37°C and shaked at 225 rpm.
3) Overnight cell culture was then centrifuged at 4000 rpm and 4°C for 10 minutes.
4) The supernatant was discarded and cell pellets were re-suspended in 5ml of Ampicilin-containing M9 medium. The amount of M9 medium was adjusted until cell suspension had an OD600 of 1.
5) The cell suspensions were then pipetted into 96-well microplate wells. This was followed by the adding of 50µl AI-2-containing supernatant into corresponding wells.
7) There were 2 different negative control samples being used for this study.
* The First control sample contained cell suspension only.
* And as the supernatants also contain large amount of LB, cell suspension with 50µl
LB added was also used as another control sample.
8) YFP measurement was carried out by VICTOR 3 multilabel reader at excitation wavelength of 490 nm and emission wavelength of 535 nm.
9) Data were automatically gathered every 10 minutes and temperature was set at 37°C.
For the control samples with only cells, the RFU remained at a steady level with time. The same thing occurred for the cells with 50µl LB broth added, although RFU level was a bit higher. For the cells with 50µl supernatants containing AI-2, we can clearly observe an increasing trend in the RFU.
This further proves that the LsrA promoter part BBa_K117002 in fact works correctly as it is activated in the presence of AI-2. Further characterization of this promoter will be implemented in future works.
From this result, it is highly expected that the detection system part BBa_K117010 also expresses lysis protein (celE7) upon induced by AI-2.
 "
"