Team:NTU-Singapore/Project
From 2008.igem.org
(Difference between revisions)
m |
(→Introduction) |
||
| (21 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <html><link rel="stylesheet" href="http:// | + | <html><link rel="stylesheet" href="http://greenbear88.googlepages.com/ntu_igem.css" type="text/css"></html> |
<div id="header">{{User:Greenbear/sandbox/header}}</div> | <div id="header">{{User:Greenbear/sandbox/header}}</div> | ||
| Line 6: | Line 6: | ||
<div id="body" style="margin-top:200px;width:950px;padding:5px;"> | <div id="body" style="margin-top:200px;width:950px;padding:5px;"> | ||
| - | + | ||
| + | |||
=Introduction= | =Introduction= | ||
| - | The focus of the NTU iGEM | + | |
| - | + | <html><b><center>ENGINEERING COLICIN E7 PRODUCTION SYSTEM TO KILL ENTEROHEMORRHAGIC ESCHERICHIA COLI O157:H7 (EHEC) VIA THE DETECTION OF EHEC SYMPTOMS </b></center><br><br> | |
| - | + | ||
| - | </ | + | |
| - | < | + | The focus of the NTU @ iGEM Team's project is the use of bacteriocins (i.e. colicin E7) for the killing of the <i>Escherichia Coli</i> O157:H7 enterohemorrhagic strain (EHEC), which causes symptoms such as bloody diarrhea and kidney failure. This is a prevalent medical problem that has a reach of a wide population. The team intends to achieve its objective by engineering a biological system that:<br><br> |
| - | </ | + | i) produces Colicin E7,<br> |
| - | <!--- | + | ii) detects the symptoms (e.g. blood) and presence of pathogenic E-Coli, and <br> |
| + | iii) releases E7 through lysis. <br> | ||
| + | <br> | ||
| + | The system will be developed using the following proposed protocol: Colicin E7 production is achieved by the insertion of the colicin E7 gene and colicin immunity gene downstream of and under regulation of the <i>lacI</i> gene, found in wild type E. Coli K12 strain. The production of the lysis protein functions under the control of an AND gate, constructed by two plasmids, and whose inputs are presence of Fe<sup>2+</sup> ions (attributed to presence of blood) and AI-2 (attributed to presence of O157:H7). <br><br> | ||
| + | |||
| + | The team's aim is to control the level of colicin production in the engineered bacteria. The engineered bacteria should also be able to be accurate in its detection. | ||
| + | |||
| + | <i>LuxS</i> knock-out was carried out to eliminate AI-2 production of K12, obtaining mutant W3110 strain, thus ensuring that AI-2 detected is purely from O157:H7. Upon detection of Fe<sup>2+</sup> ions, T7ptag expression is initiated producing T7 RNA Polymerase. Upon detection of phosphorylated AI-2, expression of the <i>supD</i> gene is initiated by activation of promoter <i>lsrA</i>, producing amber suppressor tRNA. In combination, the two upstream inputs activate the expression of the lysis protein gene, under the influence of R6K origin, resulting in amplified response of the AND gate. <br><br>The team also intends to characterize the various parts and devices used in the project, in terms of sensitivity to temperature and input concentrations by fluorescence assay and Western blotting. The team will conduct the modeling of the systems. | ||
| + | </html> | ||
| + | <br> | ||
| + | The team will employ the [https://2008.igem.org/Team:NTU-Singapore/Project/Approach methodology] derived from basic engineering concepts: Abstraction, decoupling and standardization. Please proceed to the [https://2008.igem.org/Team:NTU-Singapore/Project/Approach methodology] section for more information of our project. | ||
| + | |||
| + | |||
| + | [[Image:NTU_Comic_2.jpg|thumb|center|900px|G.O.T colicin?]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | For any comments or enquiries, feel free to contact us at [mailto:ntu.igem@gmail.com ntu.igem@gmail.com]<br><br><br> | ||
| + | <html> | ||
| + | <script language=Javascript1.2> | ||
| + | <!-- | ||
| + | |||
| + | var tags_before_clock = "<b>It is now " | ||
| + | var tags_middle_clock = "on" | ||
| + | var tags_after_clock = "</b>" | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.write('<layer id="clock"></layer><br>'); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | document.write('<span id="clock"></span>'); | ||
| + | } | ||
| + | |||
| + | DaysofWeek = new Array() | ||
| + | DaysofWeek[0]="Sunday" | ||
| + | DaysofWeek[1]="Monday" | ||
| + | DaysofWeek[2]="Tuesday" | ||
| + | DaysofWeek[3]="Wednesday" | ||
| + | DaysofWeek[4]="Thursday" | ||
| + | DaysofWeek[5]="Friday" | ||
| + | DaysofWeek[6]="Saturday" | ||
| + | |||
| + | Months = new Array() | ||
| + | Months[0]="January" | ||
| + | Months[1]="February" | ||
| + | Months[2]="March" | ||
| + | Months[3]="April" | ||
| + | Months[4]="May" | ||
| + | Months[5]="June" | ||
| + | Months[6]="July" | ||
| + | Months[7]="August" | ||
| + | Months[8]="September" | ||
| + | Months[9]="October" | ||
| + | Months[10]="November" | ||
| + | Months[11]="December" | ||
| + | |||
| + | function upclock(){ | ||
| + | var dte = new Date(); | ||
| + | var hrs = dte.getHours(); | ||
| + | var min = dte.getMinutes(); | ||
| + | var sec = dte.getSeconds(); | ||
| + | var day = DaysofWeek[dte.getDay()] | ||
| + | var date = dte.getDate() | ||
| + | var month = Months[dte.getMonth()] | ||
| + | var year = dte.getFullYear() | ||
| + | |||
| + | var col = ":"; | ||
| + | var spc = " "; | ||
| + | var com = ","; | ||
| + | var apm; | ||
| + | |||
| + | if (date == 1 || date == 21 || date == 31) | ||
| + | {ender = "<sup>st</sup>"} | ||
| + | else | ||
| + | if (date == 2 || date == 22) | ||
| + | {ender = "<sup>nd</sup>"} | ||
| + | else | ||
| + | if (date == 3 || date == 23) | ||
| + | {ender = "<sup>rd</sup>"} | ||
| + | |||
| + | else | ||
| + | {ender = "<sup>th</sup>"} | ||
| + | |||
| + | if (12 < hrs) { | ||
| + | apm="<font size='-1'>pm</font>"; | ||
| + | hrs-=12; | ||
| + | } | ||
| + | |||
| + | else { | ||
| + | apm="<font size='-1'>am</font>"; | ||
| + | } | ||
| + | |||
| + | if (hrs == 0) hrs=12; | ||
| + | if (hrs<=9) hrs="0"+hrs; | ||
| + | if (min<=9) min="0"+min; | ||
| + | if (sec<=9) sec="0"+sec; | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.clock.document.write(tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock); | ||
| + | document.clock.document.close(); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | clock.innerHTML = tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | setInterval("upclock()",1000); | ||
| + | //--> | ||
| + | </script> | ||
| + | </html> | ||
Latest revision as of 14:55, 28 October 2008
|
Introduction
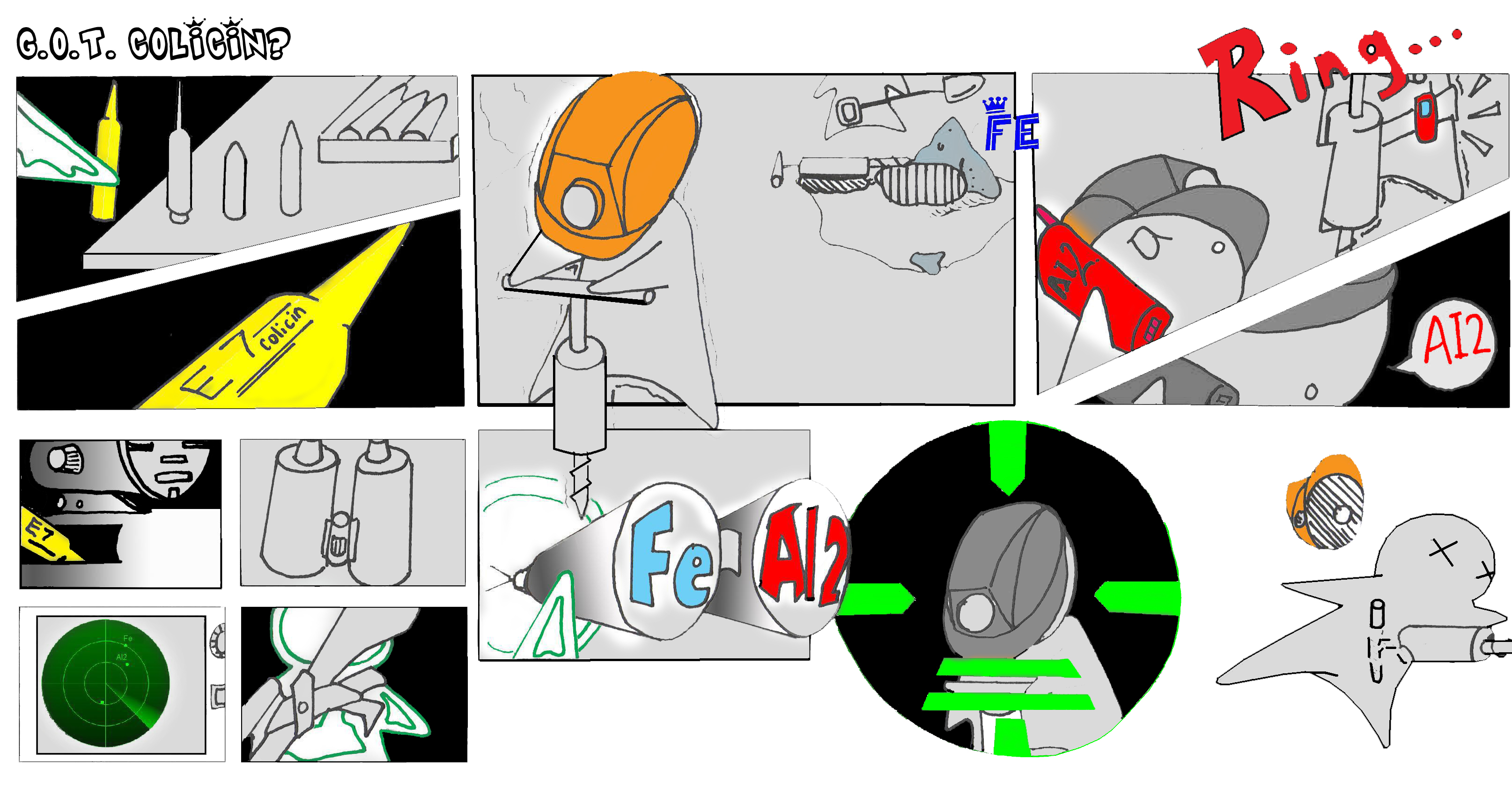
The focus of the NTU @ iGEM Team's project is the use of bacteriocins (i.e. colicin E7) for the killing of the Escherichia Coli O157:H7 enterohemorrhagic strain (EHEC), which causes symptoms such as bloody diarrhea and kidney failure. This is a prevalent medical problem that has a reach of a wide population. The team intends to achieve its objective by engineering a biological system that:
i) produces Colicin E7,
ii) detects the symptoms (e.g. blood) and presence of pathogenic E-Coli, and
iii) releases E7 through lysis.
The system will be developed using the following proposed protocol: Colicin E7 production is achieved by the insertion of the colicin E7 gene and colicin immunity gene downstream of and under regulation of the lacI gene, found in wild type E. Coli K12 strain. The production of the lysis protein functions under the control of an AND gate, constructed by two plasmids, and whose inputs are presence of Fe2+ ions (attributed to presence of blood) and AI-2 (attributed to presence of O157:H7).
The team's aim is to control the level of colicin production in the engineered bacteria. The engineered bacteria should also be able to be accurate in its detection. LuxS knock-out was carried out to eliminate AI-2 production of K12, obtaining mutant W3110 strain, thus ensuring that AI-2 detected is purely from O157:H7. Upon detection of Fe2+ ions, T7ptag expression is initiated producing T7 RNA Polymerase. Upon detection of phosphorylated AI-2, expression of the supD gene is initiated by activation of promoter lsrA, producing amber suppressor tRNA. In combination, the two upstream inputs activate the expression of the lysis protein gene, under the influence of R6K origin, resulting in amplified response of the AND gate.
The team also intends to characterize the various parts and devices used in the project, in terms of sensitivity to temperature and input concentrations by fluorescence assay and Western blotting. The team will conduct the modeling of the systems.
The team will employ the methodology derived from basic engineering concepts: Abstraction, decoupling and standardization. Please proceed to the methodology section for more information of our project.
For any comments or enquiries, feel free to contact us at ntu.igem@gmail.com
 "
"