|
|
| (25 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | Content= | | Content= |
| | <div style="font-size:18pt;"> | | <div style="font-size:18pt;"> |
| - | <font face="Arial Rounded MT Bold" style="color:#010369">__october</font></div> | + | <font face="Arial Rounded MT Bold" style="color:#010369">_october</font></div> |
| | <br> | | <br> |
| | <br> | | <br> |
| Line 139: |
Line 139: |
| | -pMA-splitlinker-Cerulan split C-CFP<br> | | -pMA-splitlinker-Cerulan split C-CFP<br> |
| | -pMA-splitlinker-Venus split C-YFP<br> | | -pMA-splitlinker-Venus split C-YFP<br> |
| - | <br> | + | <br><br>'''Detection of linkage between Fab and NIP'''(Normann)<br> |
| | + | [[Image:Freiburg08Ta50Ringer50_kont+nip.jpg]]<br> |
| | + | ''Fig: Oriogamis with (left) and without NIP given to B-cells in 50% TA-buffer(12,5mM Mg2+) and 50% Ringer (12,5mM Mg2+) ''<br><br>[[Image:Freiburg08NipnoNipinTa.jpg]]<br> |
| | + | ''Fig: Origami without (left) and with (right) NIP given to B-cells in TA-buffer (12,5mM Mg2+)''<br> Origamis seem to be absorbed by the cells. A specific linkage was not apparent.Sole TA buffer is osmotically disadvantageous for the cells. |
| | '''Analytic digestion of''' (Kathrin)<br> | | '''Analytic digestion of''' (Kathrin)<br> |
| | -transfectionvector-CMV PCR product-signalpeptide-Lipocalin -> no correct clones<br> | | -transfectionvector-CMV PCR product-signalpeptide-Lipocalin -> no correct clones<br> |
| Line 162: |
Line 165: |
| | - transfectionvector-CMV PCR product-YFP clones 1-4<br> | | - transfectionvector-CMV PCR product-YFP clones 1-4<br> |
| | - transfectionvector-CMV PCR product-Lipocalin clones 5-7<br> | | - transfectionvector-CMV PCR product-Lipocalin clones 5-7<br> |
| - | <br>
| + | |
| | test digestion of the clones named above with NotI<br> | | test digestion of the clones named above with NotI<br> |
| | analysis on the agarosegel shows not the expected bands | | analysis on the agarosegel shows not the expected bands |
| | + | <br><br> |
| | + | '''Transfection of 293t with tv-cmv-yfp'''<br> |
| | + | was done in a 6well plate <br> |
| | <br> | | <br> |
| | '''Miniprep and analytic digestion''' (Sabine)<br> | | '''Miniprep and analytic digestion''' (Sabine)<br> |
| Line 191: |
Line 197: |
| | <h3>Oct. 8th 2008</h3> <br> | | <h3>Oct. 8th 2008</h3> <br> |
| | '''Digestion of CFP''' normann<br> | | '''Digestion of CFP''' normann<br> |
| - | In order to put the gene for CFP behind the CMV-promotor on our transfectionvector to make a double trasnsfektion with YFP, the plasmit carrieng CFP was digested using pst1 and Xba1. <br> | + | In order to put the gene for CFP behind the CMV-promotor on our transfectionvector to make a double trasnsfektion with YFP, the plasmit carrying CFP was digested using pst1 and Xba1. <br> |
| | The digested gene was brought on an agarosegel and the expected band was cut out, after the followed the extraction out of the gel using the "QIAquick gel extraction kit"<br> | | The digested gene was brought on an agarosegel and the expected band was cut out, after the followed the extraction out of the gel using the "QIAquick gel extraction kit"<br> |
| | <br> | | <br> |
| Line 346: |
Line 352: |
| | <br> | | <br> |
| | <h3>Oct. 13th 2008</h3> <br> | | <h3>Oct. 13th 2008</h3> <br> |
| | + | <br> |
| | + | '''Transfection of 293t with tv-cmv-yfp''' (Normann) |
| | + | <br> done in a 6 well plate. No counted seeding this time due to too little cells in the last try.<br> |
| | '''Transformation''' (Kathrin)<br> | | '''Transformation''' (Kathrin)<br> |
| | repeat: transfectionvector-CMV PCR product clone 5+YFP clone 3 and +CFP clone 1<br> | | repeat: transfectionvector-CMV PCR product clone 5+YFP clone 3 and +CFP clone 1<br> |
| Line 411: |
Line 420: |
| | '''NIP F''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> | | '''NIP F''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> |
| | '''NIP G''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> | | '''NIP G''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> |
| - | '''NIP H''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br><br> | + | '''NIP H''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB057 (luciferase)'''<br><br> |
| | | | |
| | '''Lipo A''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> | | '''Lipo A''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> |
| Line 420: |
Line 429: |
| | '''Lipo F''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> | | '''Lipo F''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> |
| | '''Lipo G''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> | | '''Lipo G''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> |
| - | '''Lipo H''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br><br> | + | '''Lipo H''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB057 (luciferase)'''<br><br> |
| | '''2. complete parts in transfection-vector'''<br><br> | | '''2. complete parts in transfection-vector'''<br><br> |
| | '''NIP I''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> | | '''NIP I''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> |
| Line 429: |
Line 438: |
| | '''NIP VI''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> | | '''NIP VI''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> |
| | '''NIP VII''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> | | '''NIP VII''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> |
| - | '''NIP VIII''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br><br> | + | '''NIP VIII''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB057 (luciferase)'''<br><br> |
| | | | |
| | '''Lipo I''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> | | '''Lipo I''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br> |
| Line 438: |
Line 447: |
| | '''Lipo VI''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> | | '''Lipo VI''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br> |
| | '''Lipo VII''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> | | '''Lipo VII''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br> |
| - | '''Lipo VIII''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br><br> | + | '''Lipo VIII''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB057 (luciferase)'''<br><br> |
| | + | '''Transfection''' (Normann) <br> |
| | + | To test the functionality of the TV+CMV construct, a testtransfection with the construct+YFP in 293t-cells was done.<br><br> |
| | '''Miniprep''' (Kathrin)<br> | | '''Miniprep''' (Kathrin)<br> |
| | - 16ml TV_CMV_clone5_YFP clone D for transfection of 293T cells<br> | | - 16ml TV_CMV_clone5_YFP clone D for transfection of 293T cells<br> |
| Line 471: |
Line 482: |
| | <br> | | <br> |
| | '''Miniprep and analytic digestion''' (Kathrin)<br> | | '''Miniprep and analytic digestion''' (Kathrin)<br> |
| - | - pMA_transmembraneregion_Luciferase 1 <br> | + | - pMA_transmembraneregion_ß-Lactamase1 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_ß-Lactamase1 ~670bp <br> |
| - | - pMA_transmembraneregion_Luciferase 2<br> | + | - pMA_transmembraneregion_ß-Lactamase2 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_ß-Lactamase2 ~380bp<br> |
| - | - pMA_transmembraneregion_ß-Lactamase1 <br> | + | - pMA_transmembraneregion_Luciferase 1 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_Luciferase 1 ~ 350bp<br> |
| - | - pMA_transmembraneregion_ß-Lactamase2<br> | + | - pMA_transmembraneregion_Luciferase 2 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_Luciferase 2 ~ 310bp<br> |
| - | - pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker<br> | + | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) -> expected bands: pMA-Vector ~2360bp, Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) ~ 1150bp<br> |
| - | - pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F)<br>
| + | [[Image:TeamFreiburg2008_TM+bla1-bla2-BB057-BB058-lipoC_1015.jpg|250px]]<br> |
| - | - pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D)<br> | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| - | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E)<br> | + | Lane02: pMA_transmembraneregion_ß-Lactamase1 clone 1 (correct)<br> |
| - | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C)<br> | + | Lane03: pMA_transmembraneregion_ß-Lactamase1 clone 2 (correct)<br> |
| - | | + | Lane04: pMA_transmembraneregion_ß-Lactamase2 clone 1 (correct)<br> |
| - | [[Image:TeamFreiburg2008_TM+bla1-bla2-BB057-BB058-lipoC_1015.jpg|250px]] | + | Lane05: pMA_transmembraneregion_ß-Lactamase2 clone 2 (correct)<br> |
| | + | Lane06: pMA_transmembraneregion_Luciferase 1 clone 1 (correct)<br> |
| | + | Lane07: pMA_transmembraneregion_Luciferase 1 clone 2 (correct)<br> |
| | + | Lane08: pMA_transmembraneregion_Luciferase 2 clone 1 (correct)<br> |
| | + | Lane09: pMA_transmembraneregion_Luciferase 2 clone 2 (correct)<br> |
| | + | Lane10: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) clone 1 (correct)<br> |
| | + | Lane11: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) clone 2 (correct)<br> |
| | + | <br> |
| | + | - pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) -> expected bands: pMA-Vector ~2360bp, Insert Lipo D ~ 1320bp<br> |
| | + | - pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) -> expected bands: pMA-Vector ~2360bp, Insert Lipo E ~ 1140bp<br> |
| | + | - pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) -> expected bands: pMA-Vector ~2360bp, Insert Lipo F ~ 1320bp<br> |
| | + | - pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker -> expected bands: pMA-Vector ~ 2360bp, Signalpeptide-scFv-anti-NIP_GGGS-Linker ~ 920bp<br> |
| | + | - pMA-signalpeptide-scFv-anti-NIP -> expected bands: pMA-Vector ~ 2360bp, signalpeptide-Anti-Nip-sc-Fv ~ 880bp<br> |
| | + | [[Image:TeamFreiburg2008_TM+bla1-bla2-BB057-BB058-lipoC-F_1015.jpg|200px]]<br> |
| | + | Lane01: pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) clone 1 (correct)<br> |
| | + | Lane02: pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) clone 2 (correct) (correct)<br> |
| | + | Lane03: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) clone 1 (correct)<br> |
| | + | Lane04: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) clone 2 (correct)<br> |
| | + | Lane05: pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) clone 1 (correct)<br> |
| | + | Lane06: pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) clone 2 (correct)<br> |
| | + | Lane07: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 1 (correct)<br> |
| | + | Lane08: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 2 (correct)<br> |
| | + | Lane09: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 3 (correct)<br> |
| | + | Lane10: pMA-signalpeptide-scFv-anti-NIP (correct)<br> |
| | + | Lane11: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | <br> | | <br> |
| - | [[Image:TeamFreiburg2008_TM+bla1-bla2-BB057-BB058-lipoC-F_1015.jpg|200px]]
| |
| - |
| |
| - |
| |
| | '''fluorescence microscopy''' (Normann and Kathrin) <br> | | '''fluorescence microscopy''' (Normann and Kathrin) <br> |
| - | to test if the CMV-promotor in the transfectionvector allows a succesful transfection of a protein TV_CMV_5_YFP clone D was transfected to 293T cells and analysed by fluorescence microscopy. <br>
| + | To test if the CMV-promotor in the transfectionvector allows a succesful transfection of a protein TV_CMV_5_YFP clone D was transfected to 293T cells and analysed by fluorescence microscopy. <br> |
| | <br> | | <br> |
| | [[Image:Freiburg2008_150%_293Tzvi.jpg]] [[Image:Freiburg2008_Kontrolle.jpg]] [[Image:Freiburg2008_Kontrolle_Durchlicht.jpg]]<br> | | [[Image:Freiburg2008_150%_293Tzvi.jpg]] [[Image:Freiburg2008_Kontrolle.jpg]] [[Image:Freiburg2008_Kontrolle_Durchlicht.jpg]]<br> |
| | + | '''Figure_fluorescence microscopy''' from left to right: 293T cells transfected with the Transfectionvektor+CMV-promotor and a YFP construct/ control/ brightfield picture of the control<br> |
| | + | As expected the transfectionvector with CMV promotor induces expression of the yellow fluorescent protein.<br> |
| | <br> | | <br> |
| | '''Preparative Digestion''' (Sabine)<br> | | '''Preparative Digestion''' (Sabine)<br> |
| Line 516: |
Line 550: |
| | of the ligations mentioned above<br> | | of the ligations mentioned above<br> |
| | <br> | | <br> |
| - |
| |
| - | <h3>!!! New Coding for all parts !!!</h3> cause of to long descriptions<br><br>
| |
| - | '''1. complete parts in pMA-vector'''<br><br>
| |
| - | '''NIP A''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br>
| |
| - | '''NIP B''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''C-ß-lactamase'''<br>
| |
| - | '''NIP C''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-YFP'''<br>
| |
| - | '''NIP D''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-YFP'''<br>
| |
| - | '''NIP E''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-cFP'''<br>
| |
| - | '''NIP F''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br>
| |
| - | '''NIP G''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br>
| |
| - | '''NIP H''': pMA-BBFR_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br><br>
| |
| - |
| |
| - | '''Lipo A''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br>
| |
| - | '''Lipo B''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''C-ß-lactamase'''<br>
| |
| - | '''Lipo C''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-YFP'''<br>
| |
| - | '''Lipo D''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-YFP'''<br>
| |
| - | '''Lipo E''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-cFP'''<br>
| |
| - | '''Lipo F''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br>
| |
| - | '''Lipo G''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br>
| |
| - | '''Lipo H''': pMA-BBFR_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br><br>
| |
| - | '''2. complete parts in transfection-vector'''<br><br>
| |
| - | '''NIP I''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br>
| |
| - | '''NIP II''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''C-ß-lactamase'''<br>
| |
| - | '''NIP III''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-YFP'''<br>
| |
| - | '''NIP IV''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-YFP'''<br>
| |
| - | '''NIP V''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-cFP'''<br>
| |
| - | '''NIP VI''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br>
| |
| - | '''NIP VII''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br>
| |
| - | '''NIP VIII''': BBa-J52017+CMV_signalpeptide_<u>scFv-anti-NIP</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br><br>
| |
| - |
| |
| - | '''Lipo I''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-ß-lactamase'''<br>
| |
| - | '''Lipo II''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''C-ß-lactamase'''<br>
| |
| - | '''Lipo III''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-YFP'''<br>
| |
| - | '''Lipo IV''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-YFP'''<br>
| |
| - | '''Lipo V''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''SPLIT-Linker-C-cFP'''<br>
| |
| - | '''Lipo VI''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''N-CFP'''<br>
| |
| - | '''Lipo VII''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br>
| |
| - | '''Lipo VIII''': BBa-J52017+CMV_signalpeptide_<u>lipocalin</u>_GGGS-Linker_egfR-transmembraneregion_'''BB058 (luciferase)'''<br><br>
| |
| | <h3>Oct. 16th 2008</h3> <br> | | <h3>Oct. 16th 2008</h3> <br> |
| | | | |
| Line 566: |
Line 562: |
| | <h3>Oct. 17th 2008</h3> <br> | | <h3>Oct. 17th 2008</h3> <br> |
| | '''Miniprep''' (Sabine)<br> | | '''Miniprep''' (Sabine)<br> |
| - | of NIP A,B and Lipo A,B,C,D (both times clone 1 and 2)<br> | + | of NIP A,B and Lipo A,B,G,H (both times clone 1 and 2)<br> |
| | <br> | | <br> |
| | '''Analytic and Preparative Digestion''' (Sabine)<br> | | '''Analytic and Preparative Digestion''' (Sabine)<br> |
| - | of all 12 Minipreps (see above)<br>
| |
| | the analytic one with NotI, the preparative one with XbaI and PstI<br> | | the analytic one with NotI, the preparative one with XbaI and PstI<br> |
| | + | - Lipo A -> expected bands: pMA-Vector ~2360bp, Insert Lipo A ~ 1380bp<br> |
| | + | - Lipo B -> expected bands: pMA-Vector ~2360bp, Insert Lipo B ~ 1090bp<br> |
| | + | - Lipo G -> expected bands: pMA-Vector ~2360bp, Insert Lipo G ~ 1020bp<br> |
| | + | - Lipo H -> expected bands: pMA-Vector ~2360bp, Insert Lipo H ~ 1060bp<br> |
| | + | - NIP A -> expected bands: pMA-Vector ~2360bp, Insert NIP A ~ 1590bp<br> |
| | + | - NIP B -> expected bands: pMA-Vector ~2360bp, Insert NIP B ~ 1300bp<br> |
| | + | [[Image:TeamFreiburg2008_LipoA+B+G+H_NIPA+B_1017.jpg|150px]]<br> |
| | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | + | Lane02: Lipo A clone 1(correct)<br> |
| | + | Lane03: Lipo B clone 1 (correct)<br> |
| | + | Lane04: Lipo G clone 1 (correct)<br> |
| | + | Lane05: Lipo H clone 1 (correct)<br> |
| | + | Lane06: NIP A clone 1 (correct)<br> |
| | + | Lane07: NIP B clone 1 (correct)<br> |
| | <br> | | <br> |
| - | [[Image:TeamFreiburg2008_LipoA+B+G+H_NIPA+B_1017.jpg|150px]]
| |
| - |
| |
| | '''Miniprep''' (Michael)<br> | | '''Miniprep''' (Michael)<br> |
| | of NIP C,D,E,F,G and H (each with clone 1 and 2)<br><br> | | of NIP C,D,E,F,G and H (each with clone 1 and 2)<br><br> |
| | '''Analytic Digestion''' (Michael)<br> | | '''Analytic Digestion''' (Michael)<br> |
| - | of NIP C,D,E,F,G,H with NotI<br>
| + | with NotI<br> |
| | + | - NIP C -> expected bands: pMA-Vector ~2360bp, Insert NIP C ~ 1360bp<br> |
| | + | - NIP D -> expected bands: pMA-Vector ~2360bp, Insert NIP D ~ 1530bp<br> |
| | + | - NIP E -> expected bands: pMA-Vector ~2360bp, Insert NIP E ~ 1360bp<br> |
| | + | - NIP F -> expected bands: pMA-Vector ~2360bp, Insert NIP F ~ 1530bp<br> |
| | + | - NIP G -> expected bands: pMA-Vector ~2360bp, Insert NIP G ~ 1230bp<br> |
| | + | - NIP H -> expected bands: pMA-Vector ~2360bp, Insert NIP H ~ 1280bp<br> |
| | + | [[Image:TeamFreiburg2008_NIPC-H_1017.jpg|200px]]<br> |
| | + | Lane01: NIP C clone 1 (correct)<br> |
| | + | Lane02: NIP D clone 1 (correct)<br> |
| | + | Lane03: NIP E clone 1 (correct)<br> |
| | + | Lane04: NIP F clone 1 (correct)<br> |
| | + | Lane05: NIP G clone 1 (correct)<br> |
| | + | Lane06: NIP H clone 1 (correct)<br> |
| | + | Lane07: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | <br> | | <br> |
| - | [[Image:TeamFreiburg2008_NIPC-H_1017.jpg|200px]]
| |
| - |
| |
| | '''Preparative Digestion''' (Michael)<br> | | '''Preparative Digestion''' (Michael)<br> |
| | of NIP C,D,E,F,G,H with XbaI and PstI<br><br> | | of NIP C,D,E,F,G,H with XbaI and PstI<br><br> |
| Line 594: |
Line 613: |
| | <h3>Oct. 18th 2008</h3> <br> | | <h3>Oct. 18th 2008</h3> <br> |
| | '''Miniprep and analytic digestion''' (Sabine)<br> | | '''Miniprep and analytic digestion''' (Sabine)<br> |
| - | Lipo III, IV, V and VI<br> | + | - Lipo III -> expected bands: transfectionvector ~4100bp, Insert Lipo III (=Lipo C+CMV) ~ 1800bp<br> |
| | + | - Lipo IV -> expected bands: transfectionvector ~4100bp, Insert Lipo IV (=Lipo D+CMV) ~ 1970bp<br> |
| | + | - Lipo V -> expected bands: transfectionvector ~4100bp, Insert Lipo V (=Lipo E+CMV) ~ 1800bp<br> |
| | + | - Lipo VI -> expected bands: transfectionvector ~4100bp, Insert Lipo VI (=Lipo F+CMV) ~ 1970bp<br> |
| | + | [[Image:TeamFreiburg2008_Lipo3-4-5-6_1018.jpg|200px]]<br> |
| | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | + | Lane02: Lipo III clone 1 (correct)<br> |
| | + | Lane03: Lipo III clone 2 (correct)<br> |
| | + | Lane04: Lipo IV clone 1 (correct)<br> |
| | + | Lane05: Lipo IV clone 2 (correct)<br> |
| | + | Lane06: Lipo V clone 1 (not correct)<br> |
| | + | Lane07: Lipo V clone 2 (correct)<br> |
| | + | Lane08: Lipo VI clone 1 (correct))<br> |
| | + | Lane09: Lipo VI clone 2 (correct)<br> |
| | <br> | | <br> |
| - |
| |
| - | [[Image:TeamFreiburg2008_Lipo3-4-5-6_1018.jpg|200px]]
| |
| - |
| |
| | '''Ligation''' (Sabine) <br> | | '''Ligation''' (Sabine) <br> |
| | Using Quick ligase<br> | | Using Quick ligase<br> |
| Line 626: |
Line 655: |
| | <h3>Oct. 19th 2008</h3> <br> | | <h3>Oct. 19th 2008</h3> <br> |
| | '''Miniprep and analytic digestion''' (Sabine)<br> | | '''Miniprep and analytic digestion''' (Sabine)<br> |
| - | Lipo I, II, VII, VIII and NIP I, II<br> | + | - Lipo I -> expected bands: transfectionvector ~4100bp, Insert Lipo I (=Lipo A+CMV) ~ 2030bp<br> |
| | + | - Lipo II -> expected bands: transfectionvector ~4100bp, Insert Lipo II (=Lipo B+CMV) ~ 1730bp<br> |
| | + | - Lipo VII -> expected bands: transfectionvector ~4100bp, Insert Lipo VII (=Lipo G+CMV) ~ 1670bp<br> |
| | + | - Lipo VIII -> expected bands: transfectionvector ~4100bp, Insert Lipo VIII (=Lipo H+CMV) ~ 1710bp<br> |
| | + | [[Image:TeamFreiburg2008_Lipo1-2-7-8_1018.jpg|200px]]<br> |
| | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | + | Lane02: Lipo I clone 1 (correct)<br> |
| | + | Lane03: Lipo I clone 2 (correct)<br> |
| | + | Lane04: Lipo II clone 1 (correct)<br> |
| | + | Lane05: Lipo II clone 2 (correct)<br> |
| | + | Lane06: Lipo VII clone 1 (not correct)<br> |
| | + | Lane07: Lipo VII clone 2 (correct)<br> |
| | + | Lane08: Lipo VIII clone 1 (correct))<br> |
| | + | Lane09: Lipo VIII clone 2 (correct)<br> |
| | <br> | | <br> |
| - | | + | - NIP I -> expected bands: transfectionvector ~4100bp, Insert NIP I (=NIP A+CMV) ~ 2240bp<br> |
| - | [[Image:TeamFreiburg2008_Lipo1-2-7-8_1018.jpg|200px]] | + | - NIP II -> expected bands: transfectionvector ~4100bp, Insert NIP II (=NIP B+CMV) ~ 1950bp<br> |
| | + | [[Image:TeamFreiburg2008_Lipo1-2_1018.jpg|100px]]<br> |
| | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | + | Lane02: NIP I clone 1 (not correct)<br> |
| | + | Lane03: NIP I clone 2 (correct)<br> |
| | + | Lane04: NIP II clone 1 (correct)<br> |
| | + | Lane05: NIP II clone 2 (correct)<br> |
| | <br> | | <br> |
| - | [[Image:TeamFreiburg2008_Lipo1-2_1018.jpg|100px]]
| |
| - |
| |
| | '''Picking clones''' <br> | | '''Picking clones''' <br> |
| | TV/CMV-CFP, TV/CMV-YFP, NIP III, IV, V, VI, VII, VIII, pMA-transmembrane-YFP<br> | | TV/CMV-CFP, TV/CMV-YFP, NIP III, IV, V, VI, VII, VIII, pMA-transmembrane-YFP<br> |
| Line 641: |
Line 687: |
| | -TV/CMV-YFP<br> | | -TV/CMV-YFP<br> |
| | -pMA-transmembrane-YFP<br> | | -pMA-transmembrane-YFP<br> |
| - | -NIP III, IV, V, VI, VII, VIII<br> | + | -NIP III -> expected bands: transfectionvector ~4100bp, Insert NIP III (=NIP C+CMV) ~ 2010bp<br> |
| | + | -NIP IV -> expected bands: transfectionvector ~4100bp, Insert NIP IV (=NIP D+CMV) ~ 2180bp<br> |
| | + | -NIP V -> expected bands: transfectionvector ~4100bp, Insert NIP V (=NIP E+CMV) ~ 2010bp<br> |
| | + | -NIP VI -> expected bands: transfectionvector ~4100bp, Insert NIP VI (=NIP F+CMV) ~ 2180bp<br> |
| | + | -NIP VII -> expected bands: transfectionvector ~4100bp, Insert NIP VII (=NIP G+CMV) ~ 1880bp<br> |
| | + | [[Image:TeamFreiburg2008_NIPIII-VIII_1018.jpg|200px]]<br> |
| | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | + | Lane02: NIP III clone 1 (correct)<br> |
| | + | Lane03: NIP III clone 2 (correct)<br> |
| | + | Lane04: NIP IV clone 1 (not correct)<br> |
| | + | Lane05: NIP IV clone 2 (correct)<br> |
| | + | Lane06: NIP VI clone 1 (correct)<br> |
| | + | Lane07: NIP VI clone 2 (not correct)<br> |
| | + | Lane08: NIP VII clone 1 (correct))<br> |
| | + | Lane09: NIP VII clone 2 (correct)<br> |
| | <br> | | <br> |
| - | [[Image:TeamFreiburg2008_NIPIII-VIII_1018.jpg|200px]] | + | -NIP VIII -> expected bands: transfectionvector ~4100bp, Insert NIP VIII (=NIP H+CMV) ~ 1920bp<br> |
| | + | [[Image:TeamFreiburg2008_NIPVIII_1020.jpg|70px]]<br> |
| | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | + | Lane02: NIP VIII clone 1 (correct)<br> |
| | + | Lane03: NIP VIII clone 2 (correct)<br> |
| | <br> | | <br> |
| - |
| |
| - | [[Image:TeamFreiburg2008_NIPVIII_1020.jpg|70px]]
| |
| - |
| |
| | <h3>Oct. 21th 2008</h3> <br> | | <h3>Oct. 21th 2008</h3> <br> |
| | '''Sequencing''' (Kathrin)<br> | | '''Sequencing''' (Kathrin)<br> |
| Line 674: |
Line 735: |
| | -YFP, CFP, transmembrane-YFP clone 1 and 2, pMA-transmembrane, NIP I and Lipo I<br> | | -YFP, CFP, transmembrane-YFP clone 1 and 2, pMA-transmembrane, NIP I and Lipo I<br> |
| | <br> | | <br> |
| | + | '''Gelextraction of the digested constructs'''(Normann)<br>using QIAquick gel extraction kit<br><br> |
| | + | |
| | '''Miniprep''' (Kathrin)<br> | | '''Miniprep''' (Kathrin)<br> |
| | -of the 5ml cultures of the sequenced parts<br> | | -of the 5ml cultures of the sequenced parts<br> |
| | -of the 20ml cultures of the parts needed for transfection<br> | | -of the 20ml cultures of the parts needed for transfection<br> |
| | <br> | | <br> |
| - | '''Phage DNA Isolation''' (Normann)<br> | + | '''Phage DNA Isolation''' (Normann)<br>Dilution of the 50ml culture to an OD of approximately 0,1<br> |
| - | Dilution of the 50ml culture to an OD of approximately 0,1<br> | + | <br>'''Doubletransfection of 293t-cells'''(Normann) |
| | + | <br>done with TV-CMV-CFP + TV-CMV-YFP, the two parts of the anti NIP-splitbla-receptor system and the two parts of the anti NIP splitluc- receptor system. |
| | + | |
| | <br> | | <br> |
| | '''Ligation''' (Kathrin)<br> | | '''Ligation''' (Kathrin)<br> |
| Line 690: |
Line 755: |
| | Inoculate ER2738 cells with M13 phages -> shaking for approximately 4h at 37°C<br> | | Inoculate ER2738 cells with M13 phages -> shaking for approximately 4h at 37°C<br> |
| | <br> | | <br> |
| | + | After that, spinning down 50 ml at 5000g 20min and adding 7ml PEG/NaCl and storing at 4°C. Incubation over night. |
| | <h3>Oct. 23th 2008</h3> <br> | | <h3>Oct. 23th 2008</h3> <br> |
| | + | '''Detecting the new origami at the AFM (Normann)''' <br> |
| | + | [[Image:Origamis2FreiGEM08.JPG]]<br> |
| | '''Sequencing''' (Kathrin)<br> | | '''Sequencing''' (Kathrin)<br> |
| | Correct:<br> | | Correct:<br> |
| Line 696: |
Line 764: |
| | -NIP C, D and H clones 2<br> | | -NIP C, D and H clones 2<br> |
| | <br> | | <br> |
| | + | >'''Doubletransfection of 293t-cells'''(Normann) |
| | + | <br>done with TV-CMV-CFP + TV-CMV-YFP, the two parts of the anti NIP-splitbla-receptor system, the two parts of the anti NIP splitluc- receptor system and the two parts of the anti NIP split GFP receptor system.<br><br> |
| | '''Preparative Digestion''' (Kathrin)<br> | | '''Preparative Digestion''' (Kathrin)<br> |
| | NIP C, D and H (clones 2)<br> | | NIP C, D and H (clones 2)<br> |
| Line 731: |
Line 801: |
| | <h3>Oct. 24th 2008</h3> <br> | | <h3>Oct. 24th 2008</h3> <br> |
| | '''Analytic digestion''' (Sabine)<br> | | '''Analytic digestion''' (Sabine)<br> |
| - | NIP I-YFP, NIP I-CFP, Lipo I-YFP, Lipo I-CFP<br> | + | - NIP I+YFP-> expected bands: transfectionvector ~4100bp, Insert NIP I+YFP(=NIP A+CMV+YFP) ~ 2940bp<br> |
| | + | - NIP I+CFP-> expected bands: transfectionvector ~4100bp, Insert NIP I+CFP(=NIP A+CMV+CFP) ~ 2940bp<br> |
| | + | - Lipo I+YFP -> expected bands: transfectionvector ~4100bp, Insert Lipo I+YFP (=Lipo A+CMV+YFP) ~ 2730bp<br> |
| | + | - Lipo I+CFP -> expected bands: transfectionvector ~4100bp, Insert Lipo I+CFP (=Lipo A+CMV+CFP) ~ 2730bp<br> |
| | + | [[Image:TeamFreiburg2008_NIP+YFPusw_1024.jpg|250px]]<br> |
| | + | Lane01: GeneRuler 1kb DNA ladder (fermentas)<br> |
| | + | Lane02: NIP I+YFP clone 1 (correct)<br> |
| | + | Lane03: NIP I+YFP clone 2 (correct)<br> |
| | + | Lane04: NIP I+YFP clone 3 (correct)<br> |
| | + | Lane05: NIP I+CFP clone 1 (correct)<br> |
| | + | Lane06: NIP I+CFP clone 2 (correct)<br> |
| | + | Lane07: ---<br> |
| | + | Lane08: Lipo I+YFP clone 1 (correct)<br> |
| | + | Lane09: Lipo I+YFP clone 2 (correct)<br> |
| | + | Lane10: Lipo I+CFP clone 1 (correct)<br> |
| | <br> | | <br> |
| - | | + | '''staining cells with LiveBLAzer from invitrogen'''(Normann,Michael) |
| - | [[Image:TeamFreiburg2008_NIP+YFPusw_1024.jpg|250px]]
| + | <br> The cells which should have expressed both parts of the anti NIP bla receptor system were stained with LiveBLAzer, using the protocol of the invitrogen LiveBLAzer Kit.<br><br> |
| - | | + | |
| | '''Preparative digestion''' (Sabine)<br> | | '''Preparative digestion''' (Sabine)<br> |
| | -pMA-Signalpeptide-scFV-Anti-NIP-transmembrane-YFP<br> | | -pMA-Signalpeptide-scFV-Anti-NIP-transmembrane-YFP<br> |
| Line 800: |
Line 883: |
| | <br> | | <br> |
| | <h3>Oct. 27th 2008</h3> <br> | | <h3>Oct. 27th 2008</h3> <br> |
| | + | '''fluorescence microscopy''' (Michael, Sabine, Kathrin)<br> |
| | + | analysis of the transfected cells 1)-12) via fluorescence microscopy.<br> |
| | + | 1) no result <br> |
| | + | 2) no result <br> |
| | + | 3) no result <br> |
| | + | 4) no result <br> |
| | + | 5) no result <br> |
| | + | 6) no result <br> |
| | + | 7) as expected <br> |
| | + | 8) as expected <br> |
| | + | 9) as expected <br> |
| | + | 10) as expected <br> |
| | + | 11) as expected <br> |
| | + | 12) as expected <br><br> |
| | + | |
| | + | Analysis of the split beta Lactamase was performed by LiveBLAzer (invitrogen). After loading the cells with CCF4-AM the molecule is converted into CCF4 by an esterase in the cell. Beta Lactamase splits the FRET-pair CCF4 (absoption 409, emission 520nm) into two fragments. One of the fragments can still be stimulated by 409nm to detect an emission at 447nm)<br><br> |
| | + | |
| | + | [[Image:Freiburg2008_SP_LIPO_GGGS_TM_bla1_YFP 1.jpg|600px]]<br> |
| | + | <br><br> |
| | + | [[Image:Freiburg2008_SP_LIPO_GGGS_TM_bla1_YFP 2.jpg|600px]]<br> |
| | + | '''Figure_fluorescence microscopy 1''' brightfield and UV-light picture with YFP-filter. The 293T cells are transfected with the construct: Transfectionsvector+CMV_SP_Lipocalin_GGGS-linker_transmembraneregion_beta-Lactamase-fragment1_YFP.<br><br><br> |
| | + | [[Image:Freiburg2008_TV_CMV_YFP CFP_loeslich.jpg|600px]]<br> |
| | + | '''Figure_fluorescence microscopy 2''' brightfield and UV-light picture with YFP- and CFP-filter. The 293T cells are transfected with the constructs: '''upper:'''Transfectionsvector+CMV_YFP '''lower:'''Transfectionsvector+CMV_CFP<br><br> |
| | + | |
| | + | The pictures show cleary the difference between soluble YFP/CFP '''Figure_fluorescence microscopy 2''' and the membranelocalised YFP fused to the synthetic receptor construct '''Figure_fluorescence microscopy 1''' "transfectionsvector+CMV_SP_Lipocalin_GGGS-linker_transmembraneregion_beta-Lactamase-fragment1_YFP". The principle to localise our synthetic fusion-receptor-proteins to cell membrane is proved. <br> |
| | + | |
| | + | <!--<h3>Oct. 28th 2008</h3> <br>--> |
| | | | |
| | | | |
| - | <!--Bitte text über diese Zeile schreiben! -->
| |
| | }} | | }} |
|
Home
The Team
Project Report
Parts
Modeling
Notebook
Safety
CoLABoration
|
_october
Oct. 1st 2008
Digestion transfectionvector and CMV PCR product(3rd try)(Kathrin)
Digestion with EcoRI and PstI
Ligation transfectionvector and CMV PCR product(3rd try) (Kathrin)
Using new T4 DNA ligase and new ligase buffer
Miniprep of pBSK-signalpeptide (Sabine)
Transformation of pMA-transmembrane, pMA-Histag, pMA-signalpeptide, pMA-splitlinker, pMA-StreptagII (Sabine)
Oct. 2nd 2008
Picking clones (Kathrin)
pMA-transmembrane, pMA-Histag, pMA-signalpeptide, pMA-splitlinker, pMA-StreptagII
Digestion of: (Kathrin)
pBSK-signalpeptide with AgeI and PstI
pMA-Lipocalin and pMA-Anti-Nip-scFv with NgoMIV and PstI
Ligation (Kathrin)
Using Quick DNA ligase
Vector: pBSK-signalpeptide
Insert: Lipocalin, Anti-NIP-scFv
-Approaches: Insert/Vector -> a) 6/2, b) 4/4
Transformation (Kathrin)
of the ligation transfectionvector+CMV PCR product(3rd try)
Miniprep of: (Sabine)
pMA-transmembrane, pMA-Histag, pMA-signalpeptide, pMA-splitlinker, pMA-StreptagII
Preparative digestion of: (Sabine)
- pMA-splitlinker with AgeI and SpeI
- pMA-Cerulan split C-CFP and pMA-Venus split C-YFP with NgoMIV and SpeI
Ligation (Sabine)
Vector: pMA-splitlinker
Insert: Cerulan split C-CFP, Venus split C-YFP
Transformation (Sabine)
of the ligation pBSK-signalpeptide+Lipocalin and pBSK-signalpeptide+Anti-NIP-scFv
Oct. 3rd 2008
Transformation (Michael)
of the ligation pMA-splitlinker+Cerulan split C-CFP and pMA-splitlinker+Venus split C-YFP
Picking clones (Michael)
-transfectionvector-CMV PCR product
-pBSK-signalpeptide-Lipocalin
-pBSK-signalpeptide-Anti-NIP-scFv
Miniprep and analytic digestion of (Kathrin)
-transfectionvector-CMV PCR product
-pBSK-signalpeptide-Lipocalin
-pBSK-signalpeptide-Anti-NIP-scFv
Preparative digestion of (Kathrin)
-transfectionvector-CMV PCR product with SpeI and PstI
-pBSK-signalpeptide-Lipocalin and pBSK-signalpeptide-Anti-NIP-scFv with XbaI and PstI
Ligation (Sabine)
Vector: transfectionvector-CMV PCR product
Insert: signalpeptide-Lipocalin, signalpeptide-Anti-NIP-scFv
Oct. 4th 2008
Transformation (Normann)
-the ligation transfectionvector-CMV PCR product+signalpeptide-Lipocalin
-the ligation transfectionvector-CMV PCR product+signalpeptide-Anti-NIP-scFv
-pGA18-bla1(ß-Lactamase 1)
-pGA18-bla2(ß-Lactamase 2)
-pGA14-YFP
Picking clones (Sabine)
-pMA-splitlinker-Cerulan split C-CFP
-pMA-splitlinker-Venus split C-YFP
Oct. 5th 2008
Miniprep of: (Michael)
-pMA-splitlinker-Cerulan split C-CFP
-pMA-splitlinker-Venus split C-YFP
-pGA18-bla1
-pGA18-bla2
-pGA14-YFP
Picking clones (Sabine)
-transfectionvector-CMV PCR product-signalpeptide-Lipocalin
-transfectionvector-CMV PCR product-signalpeptide-Anti-NIP-scFv
Analytic digestion of: (Sabine)
-pMA-splitlinker-Cerulan split C-CFP -> expected bands: pMA-Vector ~ 2360bp, splitlinker-cCFP ~ 370bp
-pMA-splitlinker-Venus split C-YFP -> expected bands: pMA-Vector ~ 2360bp, splitlinker-cYFP ~ 370bp

Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: pMA-splitlinker-Cerulan split C-CFP clone 1 (correct)
Lane03: pMA-splitlinker-Cerulan split C-CFP clone 2 (not correct)
Lane04: pMA-splitlinker-Cerulan split C-CFP clone 3 (correct)
Lane05: pMA-splitlinker-Cerulan split C-CFP clone 4 (correct)
Lane06: ---
Lane07: pMA-splitlinker-Venus split C-YFP clone 1 (correct)
Lane08: pMA-splitlinker-Venus split C-YFP clone 2 (not correct)
Lane09: pMA-splitlinker-Venus split C-YFP clone 3 (not correct)
Lane10: pMA-splitlinker-Venus split C-YFP clone 4 (correct)
Preparative digestion (Sabine)
-pGA14-YFP
Ligation (Sabine)
Using T4 DNA ligase
Vector: transfectionvector-CMV PCR product
Insert: YFP
Oct. 6th 2008
DNA-Origamis normann
DNA-Origamis with Alexa/noNIP, NIP/Alexa, NIP, noNIP were made.
The protocol of july 24th was used.
Preparation of 293T transfection normann
In order to transfect the 293T-cells with CMV+YFP tomorrow, the amount of cells per ml was determined with the "Neubauer cell chamber" --> 2,62*10E6 cells/ml
To transfect cells with 1µg DNA in a 6-well plate, you need ~6*10E4 cells per well, so 20µl of the suspension was given in each well and 2ml DMEM was added.
Transformation of the ligation (Sabine)
transfectionvector-CMV PCR product+YFP
Preparative digestion of (Sabine)
-pMA-transmembrane with AgeI and SpeI
-pMA-Cerulan N-CFP and pMA-Venus N-YFP and NgoMIV and SpeI
Preparative digestion of (Kathrin)
-pMA-splitlinker-Cerulan split C-CFP
-pMA-splitlinker-Venus split C-YFP
Detection of linkage between Fab and NIP(Normann)

Fig: Oriogamis with (left) and without NIP given to B-cells in 50% TA-buffer(12,5mM Mg2+) and 50% Ringer (12,5mM Mg2+)
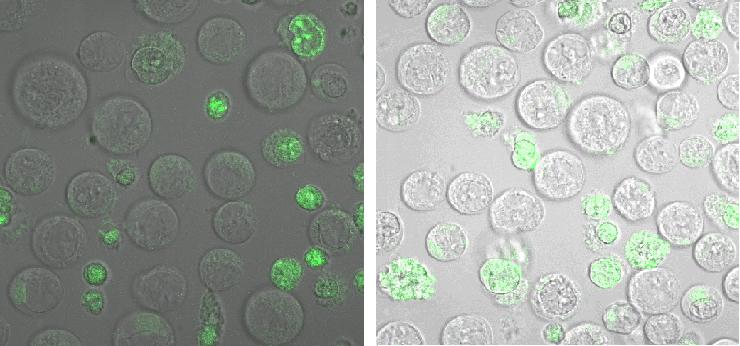
Fig: Origami without (left) and with (right) NIP given to B-cells in TA-buffer (12,5mM Mg2+)
Origamis seem to be absorbed by the cells. A specific linkage was not apparent.Sole TA buffer is osmotically disadvantageous for the cells.
Analytic digestion of (Kathrin)
-transfectionvector-CMV PCR product-signalpeptide-Lipocalin -> no correct clones
-transfectionvector-CMV PCR product-signalpeptide-Anti-NIP-scFv -> no correct clones
Ligation (Michael)
Using Quick ligase
Vector: pMA-transmembrane
Insert: splitlinker-Cerulan split C-CFP, splitlinker-Venus split C-YFP, Cerulan N-CFP and Venus N-YFP
Transformation of the ligation (Michael)
-pMA-transmembrane+splitlinker-Cerulan split C-CFP
-pMA-transmembrane+splitlinker-Venus split C-YFP
-pMA-transmembrane+Cerulan N-CFP
-pMA-transmembrane+Venus N-YFP
Picking clones (Michael)
-transfectionvector-CMV PCR product-YFP
Oct. 7th 2008
Miniprep and analytic digestion (Kathrin)
- transfectionvector-CMV PCR product-YFP clones 1-4
- transfectionvector-CMV PCR product-Lipocalin clones 5-7
test digestion of the clones named above with NotI
analysis on the agarosegel shows not the expected bands
Transfection of 293t with tv-cmv-yfp
was done in a 6well plate
Miniprep and analytic digestion (Sabine)
1) pMA-transmembrane-Cerulan N-CFP -> expected bands: pMA-Vector ~ 2360bp, transmembrane-Cerulan N-CFP ~ 610bp
2) pMA-transmembrane-Venus N-YFP -> expected bands: pMA-Vector ~ 2360bp, transmembrane-Venus N-YFP ~ 610bp
3) pMA-transmembrane-splitlinker-Cerulan split C-CFP -> expected bands: pMA-Vector ~ 2360bp, transmembrane-splitlinker-Cerulan split C-CFP ~ 430bp

Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: pMA-transmembrane-Cerulan N-CFP clone 1 (correct)
Lane03: pMA-transmembrane-Cerulan N-CFP clone 2 (correct)
Lane04: pMA-transmembrane-Cerulan N-CFP clone 3 (correct)
Lane05: pMA-transmembrane-Venus N-YFP clone 1 (correct)
Lane06: pMA-transmembrane-Venus N-YFP clone 2 (correct)
Lane07: pMA-transmembrane-Venus N-YFP clone 3 (correct)
Lane08: pMA-transmembrane-splitlinker-Cerulan split C-CFP clone 1 (correct)
Lane09: pMA-transmembrane-splitlinker-Cerulan split C-CFP clone 2 (correct)
Lane10: pMA-transmembrane-splitlinker-Cerulan split C-CFP clone 3 (correct)
4) pMA-transmembrane-splitlinker-Venus split C-YFP -> expected bands: pMA-Vector ~ 2360bp, transmembrane-splitlinker-Venus split C-YFP ~ 430bp

Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: pMA-transmembrane-splitlinker-Venus split C-YFP clone 1 (correct)
Lane03: pMA-transmembrane-splitlinker-Venus split C-YFP clone 2 (correct)
Lane04: pMA-transmembrane-splitlinker-Venus split C-YFP clone 3 (correct)
Oct. 8th 2008
Digestion of CFP normann
In order to put the gene for CFP behind the CMV-promotor on our transfectionvector to make a double trasnsfektion with YFP, the plasmit carrying CFP was digested using pst1 and Xba1.
The digested gene was brought on an agarosegel and the expected band was cut out, after the followed the extraction out of the gel using the "QIAquick gel extraction kit"
preparation of ss Phage DNA normann
Because the ss PhageDNA ,which is needed for the origamis, is nearly empty phages were harvested out of a overnight culture.
Harvesting happened like it is descriped under Methods.
Preparation of the ssDNA was done following the QIAPrep Spin M13 Kits protocol.
Klenow-fill-in-reaction (Kathrin)
For producing a GGGS-Linker
Preparative digestion (Sabine)
-pMA-transmembrane with AgeI and SpeI
-pMA-BB058(Split luciferase), pGA18-bla1, pGA18-bla2 with NgoMIV and SpeI
Ligation (Sabine)
Using Quick ligase
Vector: pMA-transmembrane
Insert: BB058(Split luciferase), bla1 and bla2
Transformation (Sabine)
of the ligations
-pMA-transmembrane+BB058(Split luciferase)
-pMA-transmembrane+bla1
-pMA-transmembrane+bla2
Miniprep (Sabine)
-transfectionvector-CMV PCR product clones 5-9
Oct. 9th 2008
Analytic digestion (with NotI) (Kathrin)
-transfectionvector-CMV PCR product clones 5-9 -> correct clones
Preparative digestion (Kathrin)
-transfectionvector-CMV PCR product clone 5 with SpeI and PstI
-pBSK-signalpeptide-Lipocalin and pBSK-signalpeptide-Anti-Nip-sc-Fv with XbaI and PstI
Sequencing (Kathrin)
Correct:
-pMA-transmembrane-Cerulan-N-CFP clone 2
-pMA-transmembrane-Venus-N-YFP clone 3
-pMA-transmembrane-splitlinker-Cerulan-C-CFP clone 2
-pMA-transmembrane-splitlinker-Venus-C-YFP clone 3
Not correct:
-pBSK-signalpeptide-Lipocalin clone 1
-pBSK-signalpeptide-Anti-NIP-sc-Fv clone 4
Analytic Digestion (Sabine)
-pMA-signalpeptide-Anti-Nip-sc-Fv -> expected bands: pMA-Vector ~2360bp, signalpeptide-Anti-Nip-sc-Fv ~880bp
-pMA-signalpeptide-Lipocalin -> expected bands: pMA-Vector ~2360bp, signalpeptide-Lipocalin ~670bp

Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: pMA-signalpeptide-Anti-Nip-sc-Fv clone 1 (correct)
Lane03: pMA-signalpeptide-Anti-Nip-sc-Fv clone 2 (correct)
Lane04: pMA-signalpeptide-Anti-Nip-sc-Fv clone 3 (correct)
Lane05: pMA-signalpeptide-Lipocalin clone 1 (correct)
Lane06: pMA-signalpeptide-Lipocalin clone 2 (correct)
Lane07: pMA-signalpeptide-Lipocalin clone 3 (correct)
Preparative Digestion (Sabine)
-pMA-BB057(Splitluciferase) with NgoMIV and SpeI
-GGGS-Linker with NgoMIV and PstI
-pMA-signalpeptide-Anti-Nip-sc-Fv clone 2 with AgeI and PstI
-pMA-signalpeptide-Lipocalin clone3 with AgeI and PstI
Ligation (Sabine)
Using T4 DNA Ligase
Vector: pMA-signalpeptide-Anti-Nip-sc-Fv, pMA-signalpeptide-Lipocalin, pMA-transmembrane
Insert: GGGS-Linker, BB057
Oct. 10th 2008
Sequencing (Kathrin)
Correct:
-pMA-signalpeptide-Anti-Nip-sc-Fv clone 2
-pMA-signalpeptide-Lipocalin clone3
Transformation (Kathrin)
of the ligations
-pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker
-pMA-signalpeptide-Lipocalin+GGGS-Linker
-pMA-transmembrane+BB057
Preparative Digestion (Sabine)
-pMA-transmembrane with AgeI and PstI
-BB057, BB058, bla1 and bla2 with NgoMIV and PstI
Picking clones (Sabine)
3 clones from pMA-signalpeptide-Lipocalin-GGGS-Linker
1 clone from pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker
Oct. 11th 2008
Ligation of CFP/YFP and Transfectionvektor+CMV
Ligation was done using the quick ligase. Inkubation for 40min.
Transformation of TV-CMV-YFP and TV-CMV-CFP
Miniprep and analytic digestion(with NotI) (Sabine)
pMA-signalpeptide-Lipocalin-GGGS-Linker clones 1-3
Ligation (Sabine)
using the Quick ligation kit
Vector: pMA-transmembrane
Insert: BB057, BB058, bla1 and bla2
Preparative Digestion (Sabine)
-pMA-signalpeptide-Lipocalin-GGGS-Linker with AgeI and PstI
-pMA-transmembrane-Cerulan-N-CFP with NgoMIV and PstI
-pMA-transmembrane-Venus-N-YFP with NgoMIV and PstI
-pMA-transmembrane-splitlinker-Cerulan-C-CFP with NgoMIV and PstI
-pMA-transmembrane-splitlinker-Venus-C-YFP with NgoMIV and PstI
-> analysis on the agarosegel showed not the expected bands
Transformation (Michael)
of the ligations
pMA-transmembrane+BB057, pMA-transmembrane+BB058, pMA-transmembrane+bla1, pMA-transmembrane+bla2
Miniprep (Michael)
pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker
Oct. 12th 2008
Miniprep (Michael)
pMA-transmembrane-Cerulan-N-CFP clone 2, pMA-transmembrane-Venus-N-YFP clone 3, pMA-transmembrane-splitlinker-Cerulan-C-CFP clone 2, pMA-transmembrane-splitlinker-Venus-C-YFP clone 3
Analytic Digestion (Kathrin)
With NotI
-pMA-signalpeptide-Anti-Nip-sc-Fv -> expected bands: pMA-Vector ~ 2360bp, pMA-signalpeptide-Anti-Nip-sc-Fv ~ 880bp
-pMA-signalpeptide-Anti-Nip-sc-Fv-GGGS-Linker -> expected bands: pMA-Vector ~ 2360bp, pMA-signalpeptide-Anti-Nip-sc-Fv-GGGSlinker ~ 920bp
-pMA-signalpeptide-Lipocalin -> expected bands: pMA-Vector ~ 2360bp, pMA-signalpeptide-Lipocalin ~ 670bp
-pMA-signalpeptide-Lipocalin-GGGS-Linker (2nd try) -> expected bands: pMA-Vector ~ 2360bp, pMA-signalpeptide-Lipocalin-GGGSlinker ~ 710bp
This time pMA-signalpeptide-Lipocalin was also digested to see the different sizes of the constructs with and without GGGS-Linker on the gel

Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: pMA-signalpeptide-Anti-Nip-sc-Fv clone 1(correct)
Lane03: pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker clone 1 (not correct)
Lane04: pMA-signalpeptide-Lipocalin clone 1(correct)
Lane05: pMA-signalpeptide-Lipocalin-GGGS-Linker clone 1 (correct)
Lane06: pMA-signalpeptide-Lipocalin-GGGS-Linker clone 2 (correct)
Lane07: pMA-signalpeptide-Lipocalin-GGGS-Linker clone 3 (correct)
Preparative Digestion (Kathrin)
-pMA-transmembrane-Cerulan-N-CFP clone 2, pMA-transmembrane-Venus-N-YFP clone 3, pMA-transmembrane-splitlinker-Cerulan-C-CFP clone 2 and pMA-transmembrane-splitlinker-Venus-C-YFP clone 3 with NgoMIV and PstI
-pMA-signalpeptide-Lipocalin-GGGS-Linker clone 3 with AgeI and PstI
-> not correct digested
-transfectionvector-CMV PCR product clone 5 with SpeI and PstI
- YFP clone 3, CFP clone 1 with XbaI and PstI
Ligation (Kathrin)
Using the Quick ligation kit
Vector: transfectionvector-CMV PCR product clone 5
Insert: YFP clone 3, CFP clone 1
Transformation (Sabine)
of the ligation transfectionvector-CMV PCR product clone 5+YFP clone 3 and transfectionvector-CMV PCR product clone 5+CFP clone 1
Oct. 13th 2008
Transfection of 293t with tv-cmv-yfp (Normann)
done in a 6 well plate. No counted seeding this time due to too little cells in the last try.
Transformation (Kathrin)
repeat: transfectionvector-CMV PCR product clone 5+YFP clone 3 and +CFP clone 1
Miniprep (Kathrin)
- pMA-transmembraneregion-Splitlinker-C-Venus-YFP clone 3
- pMA-transmembraneregion-N-Venus-YFP clone 3
- pMA-transmembraneregion-Splitlinker-C-Cerulean-CFP clone 2
- pMA-transmembraneregion-N-Cerulean-CFP clone 2
Preparative Digestion (Kathrin)
for the final construct: pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker
digestion with AgeI and PstI: pMA_Signalpeptide_scFv-anti-NIP
digestion with NgoMIV and PstI: GGGS-linker (produced by fill in reaction)
for the final construct: pMA_Signalpeptide-Lipocalin_GGGS_N-CFP/ _N-YFP/ Splitlinker_C-CFP/ Splitlinker_C-YFP
digestion with AgeI and PstI: pMA_Signalpeptide-Lipocalin_GGGS-Linker clone 1
digestion with NgoMIV and PstI: pMA_transmembraneregion_N-CFP/ _N-YFP/ Splitlinker_C-CFP/ Splitlinker_C-YFP
Analytic Digestion (Kathrin)
the following constructs were tested: Transfecionvektor_CMV_clone5_YFP A, B, C, D
after gel-seperation the expected bands were detected (3400bp=vector + 1400bp=CMV+YFP)
clone D was chosen for a transfection of 293T-cells
20ml culture of Transfecionvektor_CMV_clone5_YFP D to get enough DNA for the transfection
Preparative Digestion (Michael)
for the final construct: pMA_transmembraneregion_Luciferase1/ _Luciferase2/ -ß-Lactamase1 /- ß-Lactamase2
vector-digestion with AgeI and PstI: pMA_transmembraneregion_clone3
insert-digestion with NgoMIV and PstI: pMA_BB058 (Luciferase1), pMA_BB057 (Luciferase2), pMA_bla1 (ß-Lactamase1), pMA_bla2 (ß-Lactamase2)
Ligation (Kathrin)
for the final constructs:
- pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker
- pMA_Signalpeptide-Lipocalin_GGGS_N-CFP
- pMA_Signalpeptide-Lipocalin_GGGS_N-YFP
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP
Picking clones (Kathrin)
transfectionvector_CMV_PCR-product_clone5_YFP E, F, G, H
transfectionvector_CMV_PCR-product_clone5_CFP 1, 2, 3, 4
Ligation (Michael)
for the final constructs:
- pMA_transmembraneregion_Luciferase 1
- pMA_transmembraneregion_Luciferase 2
- pMA_transmembraneregion_ß-Lactamase1
- pMA_transmembraneregion_ß-Lactamase2
Transformation (Michael)
of all ligations mentioned above
Miniprep (Sabine)
pMA_BB057, _BB058, _transmembraneregion_clone3, _bla1, _bla2
Oct. 14th 2008
!!! New Coding for all parts !!! cause of to long descriptions
1. complete parts in pMA-vector
NIP A: pMA-BBFR_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_N-ß-lactamase
NIP B: pMA-BBFR_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_C-ß-lactamase
NIP C: pMA-BBFR_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_SPLIT-Linker-C-YFP
NIP D: pMA-BBFR_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_N-YFP
NIP E: pMA-BBFR_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_SPLIT-Linker-C-cFP
NIP F: pMA-BBFR_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_N-CFP
NIP G: pMA-BBFR_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_BB058 (luciferase)
NIP H: pMA-BBFR_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_BB057 (luciferase)
Lipo A: pMA-BBFR_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_N-ß-lactamase
Lipo B: pMA-BBFR_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_C-ß-lactamase
Lipo C: pMA-BBFR_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_SPLIT-Linker-C-YFP
Lipo D: pMA-BBFR_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_N-YFP
Lipo E: pMA-BBFR_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_SPLIT-Linker-C-cFP
Lipo F: pMA-BBFR_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_N-CFP
Lipo G: pMA-BBFR_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_BB058 (luciferase)
Lipo H: pMA-BBFR_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_BB057 (luciferase)
2. complete parts in transfection-vector
NIP I: BBa-J52017+CMV_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_N-ß-lactamase
NIP II: BBa-J52017+CMV_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_C-ß-lactamase
NIP III: BBa-J52017+CMV_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_SPLIT-Linker-C-YFP
NIP IV: BBa-J52017+CMV_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_N-YFP
NIP V: BBa-J52017+CMV_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_SPLIT-Linker-C-cFP
NIP VI: BBa-J52017+CMV_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_N-CFP
NIP VII: BBa-J52017+CMV_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_BB058 (luciferase)
NIP VIII: BBa-J52017+CMV_signalpeptide_scFv-anti-NIP_GGGS-Linker_egfR-transmembraneregion_BB057 (luciferase)
Lipo I: BBa-J52017+CMV_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_N-ß-lactamase
Lipo II: BBa-J52017+CMV_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_C-ß-lactamase
Lipo III: BBa-J52017+CMV_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_SPLIT-Linker-C-YFP
Lipo IV: BBa-J52017+CMV_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_N-YFP
Lipo V: BBa-J52017+CMV_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_SPLIT-Linker-C-cFP
Lipo VI: BBa-J52017+CMV_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_N-CFP
Lipo VII: BBa-J52017+CMV_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_BB058 (luciferase)
Lipo VIII: BBa-J52017+CMV_signalpeptide_lipocalin_GGGS-Linker_egfR-transmembraneregion_BB057 (luciferase)
Transfection (Normann)
To test the functionality of the TV+CMV construct, a testtransfection with the construct+YFP in 293t-cells was done.
Miniprep (Kathrin)
- 16ml TV_CMV_clone5_YFP clone D for transfection of 293T cells
- 4ml TV_CMV_clone5_CFP clone 1, 2, 3, 4
- 4ml TV_CMV_clone5_YFP clone E, F, G, H
of the 4ml cultures also glycerinstockes were taken
Analytic Digestion (Kathrin)
- TV_CMV_clone5_CFP clone 1, 2, 3, 4
- TV_CMV_clone5_YFP clone E, F, G, H
expected bands were detected (3400bp=vector + 1400bp=CMV+YFP/ CMV+CFP)
Sequencing (Kathrin)
Correct:
- pMA_Signalpeptide_Lipocalin_GGGS-linker_clone 1
- pMA_Signalpeptide_Lipocalin_GGGS-linker_clone 2
Not correct
- TV_CMV_clone5_YFP clone A (base exchange in the CMV-promotor)
Picking clones (Michael)
- pMA_transmembraneregion_Luciferase 1
- pMA_transmembraneregion_Luciferase 2
- pMA_transmembraneregion_ß-Lactamase1
- pMA_transmembraneregion_ß-Lactamase2
- pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker
- pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F)
- pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D)
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E)
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C)
Oct. 15th 2008
Miniprep and analytic digestion (Kathrin)
- pMA_transmembraneregion_ß-Lactamase1 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_ß-Lactamase1 ~670bp
- pMA_transmembraneregion_ß-Lactamase2 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_ß-Lactamase2 ~380bp
- pMA_transmembraneregion_Luciferase 1 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_Luciferase 1 ~ 350bp
- pMA_transmembraneregion_Luciferase 2 -> expected bands: pMA-Vector ~2360bp, transmembraneregion_Luciferase 2 ~ 310bp
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) -> expected bands: pMA-Vector ~2360bp, Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) ~ 1150bp

Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: pMA_transmembraneregion_ß-Lactamase1 clone 1 (correct)
Lane03: pMA_transmembraneregion_ß-Lactamase1 clone 2 (correct)
Lane04: pMA_transmembraneregion_ß-Lactamase2 clone 1 (correct)
Lane05: pMA_transmembraneregion_ß-Lactamase2 clone 2 (correct)
Lane06: pMA_transmembraneregion_Luciferase 1 clone 1 (correct)
Lane07: pMA_transmembraneregion_Luciferase 1 clone 2 (correct)
Lane08: pMA_transmembraneregion_Luciferase 2 clone 1 (correct)
Lane09: pMA_transmembraneregion_Luciferase 2 clone 2 (correct)
Lane10: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) clone 1 (correct)
Lane11: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP (Lipo C) clone 2 (correct)
- pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) -> expected bands: pMA-Vector ~2360bp, Insert Lipo D ~ 1320bp
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) -> expected bands: pMA-Vector ~2360bp, Insert Lipo E ~ 1140bp
- pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) -> expected bands: pMA-Vector ~2360bp, Insert Lipo F ~ 1320bp
- pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker -> expected bands: pMA-Vector ~ 2360bp, Signalpeptide-scFv-anti-NIP_GGGS-Linker ~ 920bp
- pMA-signalpeptide-scFv-anti-NIP -> expected bands: pMA-Vector ~ 2360bp, signalpeptide-Anti-Nip-sc-Fv ~ 880bp

Lane01: pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) clone 1 (correct)
Lane02: pMA_Signalpeptide-Lipocalin_GGGS_N-YFP (Lipo D) clone 2 (correct) (correct)
Lane03: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) clone 1 (correct)
Lane04: pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP (Lipo E) clone 2 (correct)
Lane05: pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) clone 1 (correct)
Lane06: pMA_Signalpeptide-Lipocalin_GGGS_N-CFP (Lipo F) clone 2 (correct)
Lane07: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 1 (correct)
Lane08: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 2 (correct)
Lane09: pMA_Signalpeptide-scFv-anti-NIP_GGGS-Linker clone 3 (correct)
Lane10: pMA-signalpeptide-scFv-anti-NIP (correct)
Lane11: GeneRuler 1kb DNA ladder (fermentas)
fluorescence microscopy (Normann and Kathrin)
To test if the CMV-promotor in the transfectionvector allows a succesful transfection of a protein TV_CMV_5_YFP clone D was transfected to 293T cells and analysed by fluorescence microscopy.
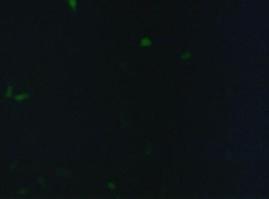  
Figure_fluorescence microscopy from left to right: 293T cells transfected with the Transfectionvektor+CMV-promotor and a YFP construct/ control/ brightfield picture of the control
As expected the transfectionvector with CMV promotor induces expression of the yellow fluorescent protein.
Preparative Digestion (Sabine)
for the final constructs: pMA_signalpeptide_scFv-antiNIP_GGGS-linker_transmembraneregion_bla1/ _bla2/ _Luciferase-BB058/ _Luciferase-BB057
digestion with AgeI and PstI: pMA_signalpeptide_scFv-antiNIP_GGGS-linker_
digestion with NgoMIV and PstI:pMA_transmembraneregion_bla1/ _bla2/ _Luciferase-BB058/ _Luciferase-BB057
Sequencing (Kathrin)
Correct:
- TV_CMV_clone 7
- TV_CMV_clone 8
Not correct
- TV_CMV_clone 5 (base exchange in the CMV promotor)
- TV_CMV_clone 9 (base exchange in the CMV promotor)
- TV_CMV_clone5_YFP A (base exchange in the CMV promotor)
- TV_CMV_clone5_CFP 1 (base exchange in the CMV promotor)
Ligation (Michael)
Using Quick ligase
Vector: pMA-signalpeptide-scFV-antiNIP-GGGSlinker
Insert: transmembrane-bla1, transmembrane-bla2, transmembrane-splitlinker-C-CFP, transmembrane-splitlinker-C-YFP, transmemrbane-N-CFP, transmembrane-N-YFP, transmembrane-Splitluciferase(BB058), transmembrane-Splitluciferase(BB057)
Vector: pMA-signalpeptide-Lipocalin-GGGSlinker
Insert: transmembrane-bla1, transmembrane-bla2, transmembrane-Splitluciferase(BB058), transmembrane-Splitluciferase(BB057)
Transformation (Michael)
of the ligations mentioned above
Oct. 16th 2008
Preparative Digestion (Sabine)
of Lipo C,D,E and F (always clone 1) with XbaI and PstI
to bring into the transfection-vector TV/CMV clone 7, digested with SpeI and PstI
Ligation (Michael)
of the digested vector and inserts (see above) with Quick ligase
Transformation (Michael)
of Lipo III, IV, V and VI in XL-1 cells
Oct. 17th 2008
Miniprep (Sabine)
of NIP A,B and Lipo A,B,G,H (both times clone 1 and 2)
Analytic and Preparative Digestion (Sabine)
the analytic one with NotI, the preparative one with XbaI and PstI
- Lipo A -> expected bands: pMA-Vector ~2360bp, Insert Lipo A ~ 1380bp
- Lipo B -> expected bands: pMA-Vector ~2360bp, Insert Lipo B ~ 1090bp
- Lipo G -> expected bands: pMA-Vector ~2360bp, Insert Lipo G ~ 1020bp
- Lipo H -> expected bands: pMA-Vector ~2360bp, Insert Lipo H ~ 1060bp
- NIP A -> expected bands: pMA-Vector ~2360bp, Insert NIP A ~ 1590bp
- NIP B -> expected bands: pMA-Vector ~2360bp, Insert NIP B ~ 1300bp

Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: Lipo A clone 1(correct)
Lane03: Lipo B clone 1 (correct)
Lane04: Lipo G clone 1 (correct)
Lane05: Lipo H clone 1 (correct)
Lane06: NIP A clone 1 (correct)
Lane07: NIP B clone 1 (correct)
Miniprep (Michael)
of NIP C,D,E,F,G and H (each with clone 1 and 2)
Analytic Digestion (Michael)
with NotI
- NIP C -> expected bands: pMA-Vector ~2360bp, Insert NIP C ~ 1360bp
- NIP D -> expected bands: pMA-Vector ~2360bp, Insert NIP D ~ 1530bp
- NIP E -> expected bands: pMA-Vector ~2360bp, Insert NIP E ~ 1360bp
- NIP F -> expected bands: pMA-Vector ~2360bp, Insert NIP F ~ 1530bp
- NIP G -> expected bands: pMA-Vector ~2360bp, Insert NIP G ~ 1230bp
- NIP H -> expected bands: pMA-Vector ~2360bp, Insert NIP H ~ 1280bp

Lane01: NIP C clone 1 (correct)
Lane02: NIP D clone 1 (correct)
Lane03: NIP E clone 1 (correct)
Lane04: NIP F clone 1 (correct)
Lane05: NIP G clone 1 (correct)
Lane06: NIP H clone 1 (correct)
Lane07: GeneRuler 1kb DNA ladder (fermentas)
Preparative Digestion (Michael)
of NIP C,D,E,F,G,H with XbaI and PstI
Picking clones (Sabine)
Lipo III, IV, V and VI
Ligation (Sabine)
Inserts: NIP C, D, E, F, G, H (clone 1)
Vector: TV/CMV (clon 7)
with ligase T4
Miniprep (Sabine)
of Lipo III, IV, V and VI
Oct. 18th 2008
Miniprep and analytic digestion (Sabine)
- Lipo III -> expected bands: transfectionvector ~4100bp, Insert Lipo III (=Lipo C+CMV) ~ 1800bp
- Lipo IV -> expected bands: transfectionvector ~4100bp, Insert Lipo IV (=Lipo D+CMV) ~ 1970bp
- Lipo V -> expected bands: transfectionvector ~4100bp, Insert Lipo V (=Lipo E+CMV) ~ 1800bp
- Lipo VI -> expected bands: transfectionvector ~4100bp, Insert Lipo VI (=Lipo F+CMV) ~ 1970bp
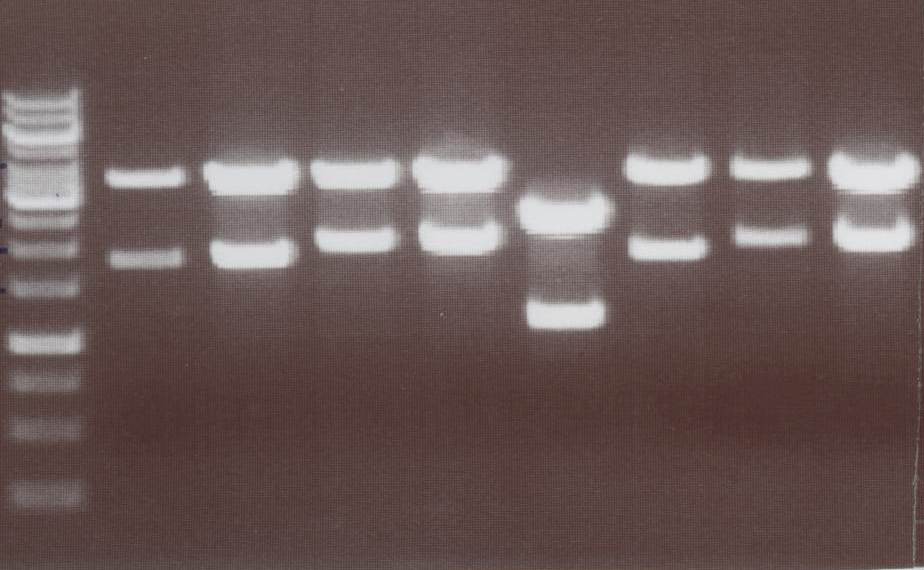
Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: Lipo III clone 1 (correct)
Lane03: Lipo III clone 2 (correct)
Lane04: Lipo IV clone 1 (correct)
Lane05: Lipo IV clone 2 (correct)
Lane06: Lipo V clone 1 (not correct)
Lane07: Lipo V clone 2 (correct)
Lane08: Lipo VI clone 1 (correct))
Lane09: Lipo VI clone 2 (correct)
Ligation (Sabine)
Using Quick ligase
Vector: transfection-vector TV/CMV clone 7
Insert: CFP clone 1, YFP clone 3
Preparative digestion (Sabine)
-pMA-transmembrane with AgeI and PstI
-YFP clone 3 with NgoMIV and PstI
Transformation (Sabine)
of the quick ligations
-transfection-vector TV/CMV clone 7 + CFP clone 1
-transfection-vector TV/CMV clone 7 + YFP clone 3
of the ligations with T4 ligase (Oct. 17th 2008)
-transfection-vector TV/CMV clone 7 + NIP C, D, E, F, G, H
of the part BCR-transmembrane ordered by geneart
Picking clones (Sabine)
Lipo I, II, VII, VIII and NIP I, II
Ligation and transformation (Michael)
Using Quick ligase
Vector: pMA-transmembrane
Insert: YFP
Oct. 19th 2008
Miniprep and analytic digestion (Sabine)
- Lipo I -> expected bands: transfectionvector ~4100bp, Insert Lipo I (=Lipo A+CMV) ~ 2030bp
- Lipo II -> expected bands: transfectionvector ~4100bp, Insert Lipo II (=Lipo B+CMV) ~ 1730bp
- Lipo VII -> expected bands: transfectionvector ~4100bp, Insert Lipo VII (=Lipo G+CMV) ~ 1670bp
- Lipo VIII -> expected bands: transfectionvector ~4100bp, Insert Lipo VIII (=Lipo H+CMV) ~ 1710bp
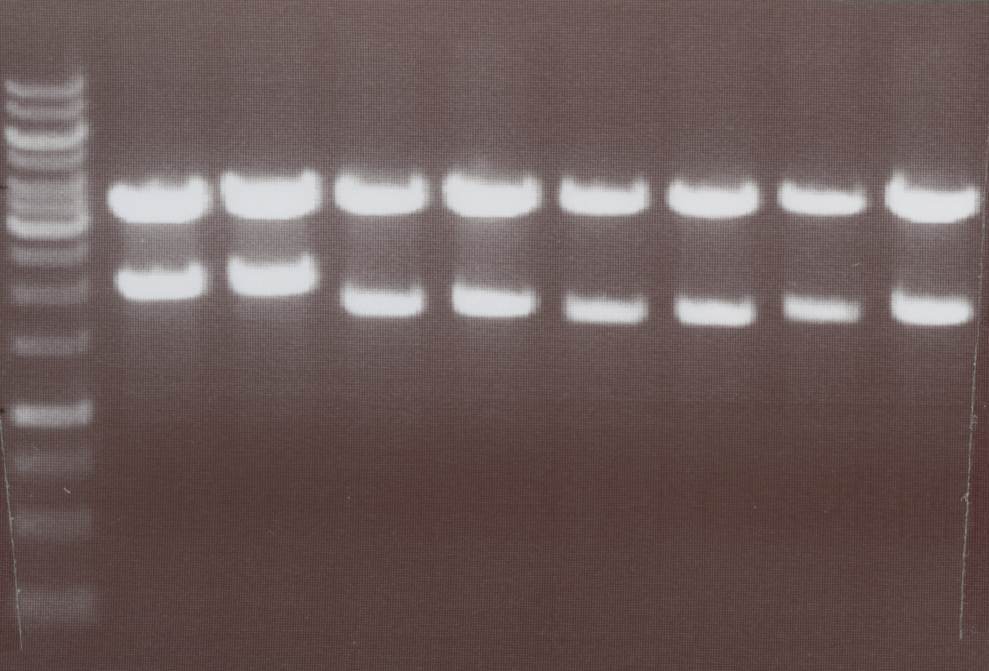
Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: Lipo I clone 1 (correct)
Lane03: Lipo I clone 2 (correct)
Lane04: Lipo II clone 1 (correct)
Lane05: Lipo II clone 2 (correct)
Lane06: Lipo VII clone 1 (not correct)
Lane07: Lipo VII clone 2 (correct)
Lane08: Lipo VIII clone 1 (correct))
Lane09: Lipo VIII clone 2 (correct)
- NIP I -> expected bands: transfectionvector ~4100bp, Insert NIP I (=NIP A+CMV) ~ 2240bp
- NIP II -> expected bands: transfectionvector ~4100bp, Insert NIP II (=NIP B+CMV) ~ 1950bp
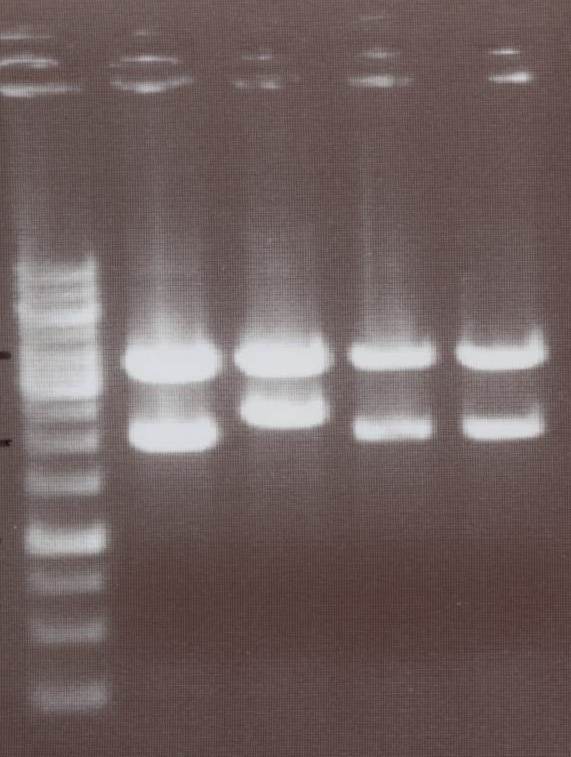
Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: NIP I clone 1 (not correct)
Lane03: NIP I clone 2 (correct)
Lane04: NIP II clone 1 (correct)
Lane05: NIP II clone 2 (correct)
Picking clones
TV/CMV-CFP, TV/CMV-YFP, NIP III, IV, V, VI, VII, VIII, pMA-transmembrane-YFP
Oct. 20th 2008
Miniprep and analytic digestion (Michael)
-TV/CMV-CFP
-TV/CMV-YFP
-pMA-transmembrane-YFP
-NIP III -> expected bands: transfectionvector ~4100bp, Insert NIP III (=NIP C+CMV) ~ 2010bp
-NIP IV -> expected bands: transfectionvector ~4100bp, Insert NIP IV (=NIP D+CMV) ~ 2180bp
-NIP V -> expected bands: transfectionvector ~4100bp, Insert NIP V (=NIP E+CMV) ~ 2010bp
-NIP VI -> expected bands: transfectionvector ~4100bp, Insert NIP VI (=NIP F+CMV) ~ 2180bp
-NIP VII -> expected bands: transfectionvector ~4100bp, Insert NIP VII (=NIP G+CMV) ~ 1880bp
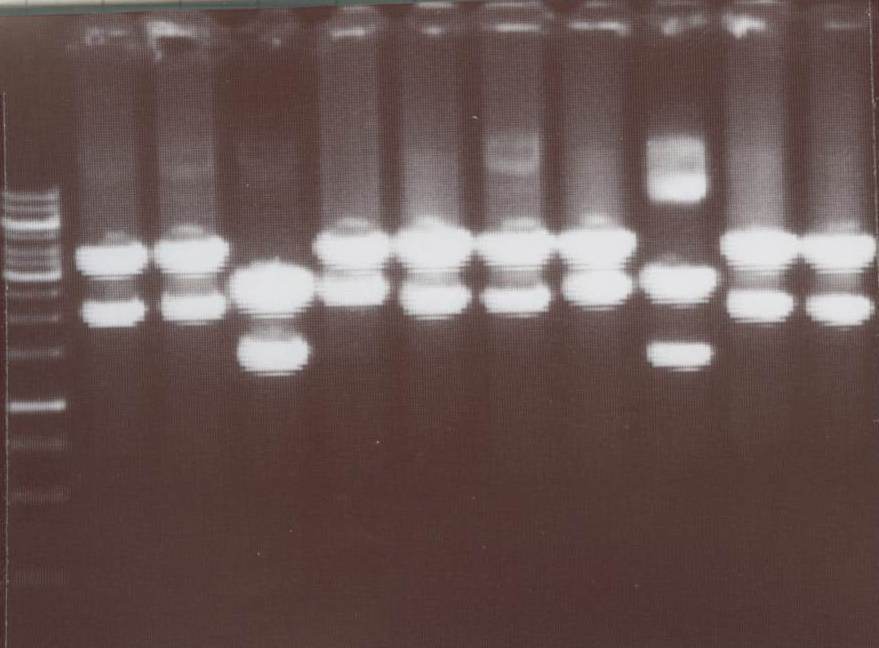
Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: NIP III clone 1 (correct)
Lane03: NIP III clone 2 (correct)
Lane04: NIP IV clone 1 (not correct)
Lane05: NIP IV clone 2 (correct)
Lane06: NIP VI clone 1 (correct)
Lane07: NIP VI clone 2 (not correct)
Lane08: NIP VII clone 1 (correct))
Lane09: NIP VII clone 2 (correct)
-NIP VIII -> expected bands: transfectionvector ~4100bp, Insert NIP VIII (=NIP H+CMV) ~ 1920bp
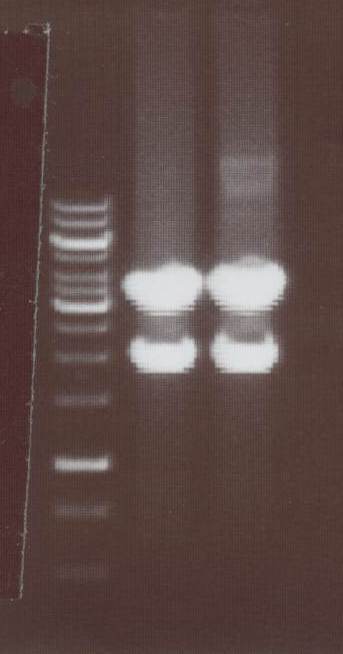
Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: NIP VIII clone 1 (correct)
Lane03: NIP VIII clone 2 (correct)
Oct. 21th 2008
Sequencing (Kathrin)
Correct:
-pMA-signalpeptide-scFV-Anti-NIP-GGGSlinker clone 1 and clone 2
-pMA-transmembrane-BB058(Luciferase) clone 1
-pMA-transmembrane-BB057(Luciferase) clone 1
-NIP A, B, E, F, and G
-Lipo A, B, C, D, E, F, G and H
Not correct(base exchanges):
-NIP C, D and H (clones 1)
5ml cultures (Kathrin)
of the sequenced parts
Phage DNA Isolation (Normann)
50ml culture of ER2738 cells
Seeding 293T cells for transfection (Normann)
20ml cultures for transfection (Kathrin)
cultures of the parts using for transfection (NipI-NipVIII, TV/CMV-CFP, TV/CMV-YFP)
Oct. 22th 2008
Preparative Digestion (Kathrin)
-YFP, CFP, transmembrane-YFP clone 1 and 2, pMA-transmembrane, NIP I and Lipo I
Gelextraction of the digested constructs(Normann)
using QIAquick gel extraction kit
Miniprep (Kathrin)
-of the 5ml cultures of the sequenced parts
-of the 20ml cultures of the parts needed for transfection
Phage DNA Isolation (Normann)
Dilution of the 50ml culture to an OD of approximately 0,1
Doubletransfection of 293t-cells(Normann)
done with TV-CMV-CFP + TV-CMV-YFP, the two parts of the anti NIP-splitbla-receptor system and the two parts of the anti NIP splitluc- receptor system.
Ligation (Kathrin)
-Vector: pMA-transmembrane, Insert: YFP, CFP
-Vector: pMA-signalpeptide-scFV-Anti-NIP-GGGS, Insert: transmembrane-YFP, transmembrane-CFP
-Vector: pMA-signalpeptide-Lipocalin-GGGS, Insert: transmembrane-YFP, transmembrane-CFP
-Vector: Lipo I, Insert: YFP, CFP
-Vector: NIP I, Insert: YFP, CFP
Phage DNA Isolation (Normann)
Inoculate ER2738 cells with M13 phages -> shaking for approximately 4h at 37°C
After that, spinning down 50 ml at 5000g 20min and adding 7ml PEG/NaCl and storing at 4°C. Incubation over night.
Oct. 23th 2008
Detecting the new origami at the AFM (Normann)

Sequencing (Kathrin)
Correct:
-TV/CMV-CFP clone 1
-NIP C, D and H clones 2
>Doubletransfection of 293t-cells(Normann)
done with TV-CMV-CFP + TV-CMV-YFP, the two parts of the anti NIP-splitbla-receptor system, the two parts of the anti NIP splitluc- receptor system and the two parts of the anti NIP split GFP receptor system.
Preparative Digestion (Kathrin)
NIP C, D and H (clones 2)
Ligation (Kathrin)
Using Quick ligase
Vector: TV/CMV
Insert: NIP C, D and H (clones 2)
Transformation (Michael
of the ligations (see above)
5ml cultures (Kathrin)
-NIP C, D and H (clones 2)
-pMA-signalpeptide-NIP-GGGS clone 1 and 2
-pMA-transmembrane-betaLactamase1 clone 1
-pMA-transmembrane-betaLactamase2 clone 1
-pMA-transmembrane-BB058(Splitluciferase) clone 1
-pMA-transmembrane-BB057(Splitluciferase) clone 1
Miniprep (Sabine)
-NIP C, D and H (clones 2)
-pMA-signalpeptide-NIP-GGGS clone 1 and 2
-pMA-transmembrane-betaLactamase1 clone 1
-pMA-transmembrane-betaLactamase2 clone 1
-pMA-transmembrane-BB058(Splitluciferase) clone 1
-pMA-transmembrane-BB057(Splitluciferase) clone 1
Ligation(2nd try) (Simone)
Using Quick ligase
Vector: TV/CMV
Insert: NIP C, D and H (clones 2)
Transformation (Sabine)
of the ligations (see above)
Oct. 24th 2008
Analytic digestion (Sabine)
- NIP I+YFP-> expected bands: transfectionvector ~4100bp, Insert NIP I+YFP(=NIP A+CMV+YFP) ~ 2940bp
- NIP I+CFP-> expected bands: transfectionvector ~4100bp, Insert NIP I+CFP(=NIP A+CMV+CFP) ~ 2940bp
- Lipo I+YFP -> expected bands: transfectionvector ~4100bp, Insert Lipo I+YFP (=Lipo A+CMV+YFP) ~ 2730bp
- Lipo I+CFP -> expected bands: transfectionvector ~4100bp, Insert Lipo I+CFP (=Lipo A+CMV+CFP) ~ 2730bp

Lane01: GeneRuler 1kb DNA ladder (fermentas)
Lane02: NIP I+YFP clone 1 (correct)
Lane03: NIP I+YFP clone 2 (correct)
Lane04: NIP I+YFP clone 3 (correct)
Lane05: NIP I+CFP clone 1 (correct)
Lane06: NIP I+CFP clone 2 (correct)
Lane07: ---
Lane08: Lipo I+YFP clone 1 (correct)
Lane09: Lipo I+YFP clone 2 (correct)
Lane10: Lipo I+CFP clone 1 (correct)
staining cells with LiveBLAzer from invitrogen(Normann,Michael)
The cells which should have expressed both parts of the anti NIP bla receptor system were stained with LiveBLAzer, using the protocol of the invitrogen LiveBLAzer Kit.
Preparative digestion (Sabine)
-pMA-Signalpeptide-scFV-Anti-NIP-transmembrane-YFP
-pMA-Signalpeptide-Lipocalin-transmembrane-YFP
Miniprep (Kathrin)
-pMA-transmembrane-YFP and pMA-transmembrane-CFP
-NIP I-YFP, NIP I-CFP, Lipo I-YFP, Lipo I-CFP
Preparative digestion (Kathrin)
pMA-transmembrane-YFP and pMA-transmembrane-CFP with NgoMIV and PstI
Ligation and transformation (Sabine)
Using Quick ligase
-Vector: pMA-signalpeptide-scFv-Anti-NIP-GGGS, Insert: transmembrane-YFP, transmembrane-CFP
-Vector: TV/CMV Insert: scFv-Anti-NIP-transmembrane-YFP
Oct. 25th 2008
Miniprep and analytic digestion (Sabine)
NIP III, NIP IV and NIP VIII
-> NIP IV showed not the expected bands -> new clones were picked (clones 4-7)
Ligation and transformation(2nd try) (Sabine)
Using Quick ligase
-Vector: pMA-signalpeptide-scFv-Anti-NIP-GGGS, Insert: transmembrane-YFP, transmembrane-CFP
-Vector: TV/CMV Insert: scFv-Anti-NIP-transmembrane-YFP
Seeding 293T cells for transfection (Sabine)
Oct. 26th 2008
Miniprep and analytic digestion (Kathrin)
-NIP IV (clones 4-7)
-pMA-signalpeptide-scFv-anti-NIP-GGGS-transmembrane-CFP
Preparative digestion (Kathrin)
-pMA-signalpeptide-scFV-anti-NIP-transmembrane-CFP with XbaI and PstI
-TV/CMV with SpeI and PstI
Picking clones (Kathrin)
-pMA-signalpeptide-scFv-Anti-NIP-transmembrane-YFP, pMA-signalpeptide-scFv-Anti-NIP-transmembrane-CFP
-transfectionvector-Lipocalin-transmembrane-YFP, transfectionvector-scFV-Anti_NIP-transmembrane-YFP
Ligation (Sabine)
-Vector: TV/CMV, Insert: NIP D clone 2 (2nd try, since the analytic digestion of several clones showed not the expected bands)
-Vector: TV/CMV, Insert: scFV-Anti-NIP-GGGS-transmembrane-CFP, scFV-Anti-NIP-GGGS-transmembrane-YFP
Transformation (Michael)
of the ligations (see above)
Transfection of 293T cells (Sabine)
For transfection 2µg DNA (4µg DNA for double transfection) was used
1) Double transfection with NIP I and NIP II
2) Double transfection with NIP III and NIP IV
3) Double transfection with NIP V and NIP VI
4) Double transfection with NIP VII and NIP VIII
5) NIP I-YFP
6) NIP I-CFP
7) Lipo I-YFP
8) Lipo I-CFP
9) TV/CMV-YFP 1
10)TV/CMV-CFP
11)TV/CMV-YFP A
12)Negative control
Oct. 27th 2008
fluorescence microscopy (Michael, Sabine, Kathrin)
analysis of the transfected cells 1)-12) via fluorescence microscopy.
1) no result
2) no result
3) no result
4) no result
5) no result
6) no result
7) as expected
8) as expected
9) as expected
10) as expected
11) as expected
12) as expected
Analysis of the split beta Lactamase was performed by LiveBLAzer (invitrogen). After loading the cells with CCF4-AM the molecule is converted into CCF4 by an esterase in the cell. Beta Lactamase splits the FRET-pair CCF4 (absoption 409, emission 520nm) into two fragments. One of the fragments can still be stimulated by 409nm to detect an emission at 447nm)
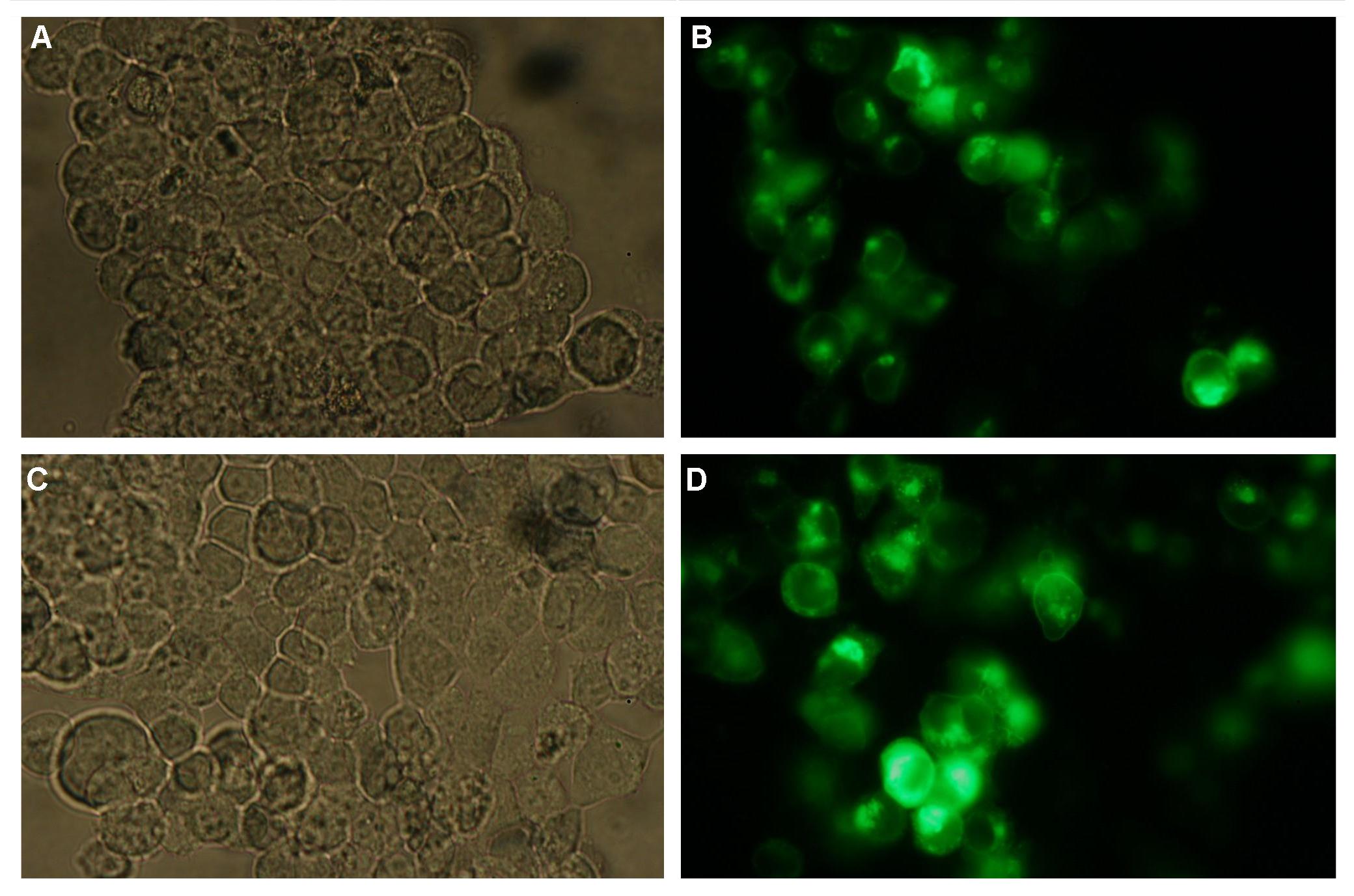
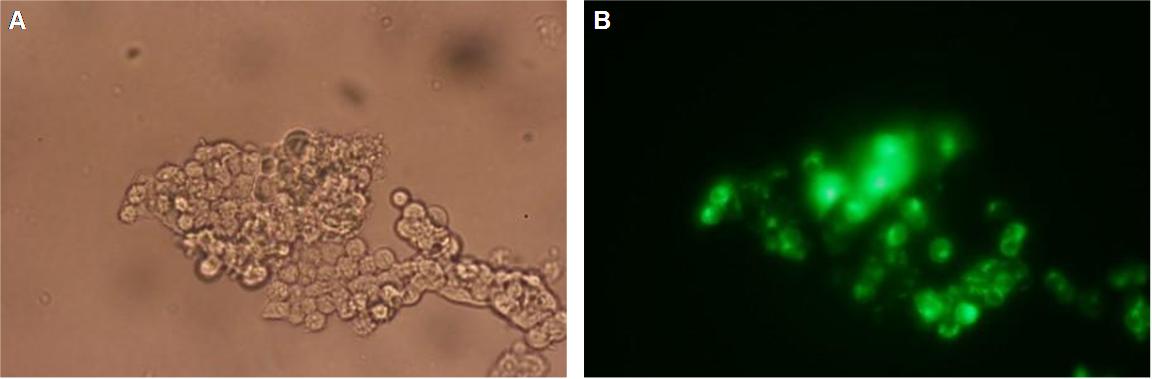
Figure_fluorescence microscopy 1 brightfield and UV-light picture with YFP-filter. The 293T cells are transfected with the construct: Transfectionsvector+CMV_SP_Lipocalin_GGGS-linker_transmembraneregion_beta-Lactamase-fragment1_YFP.
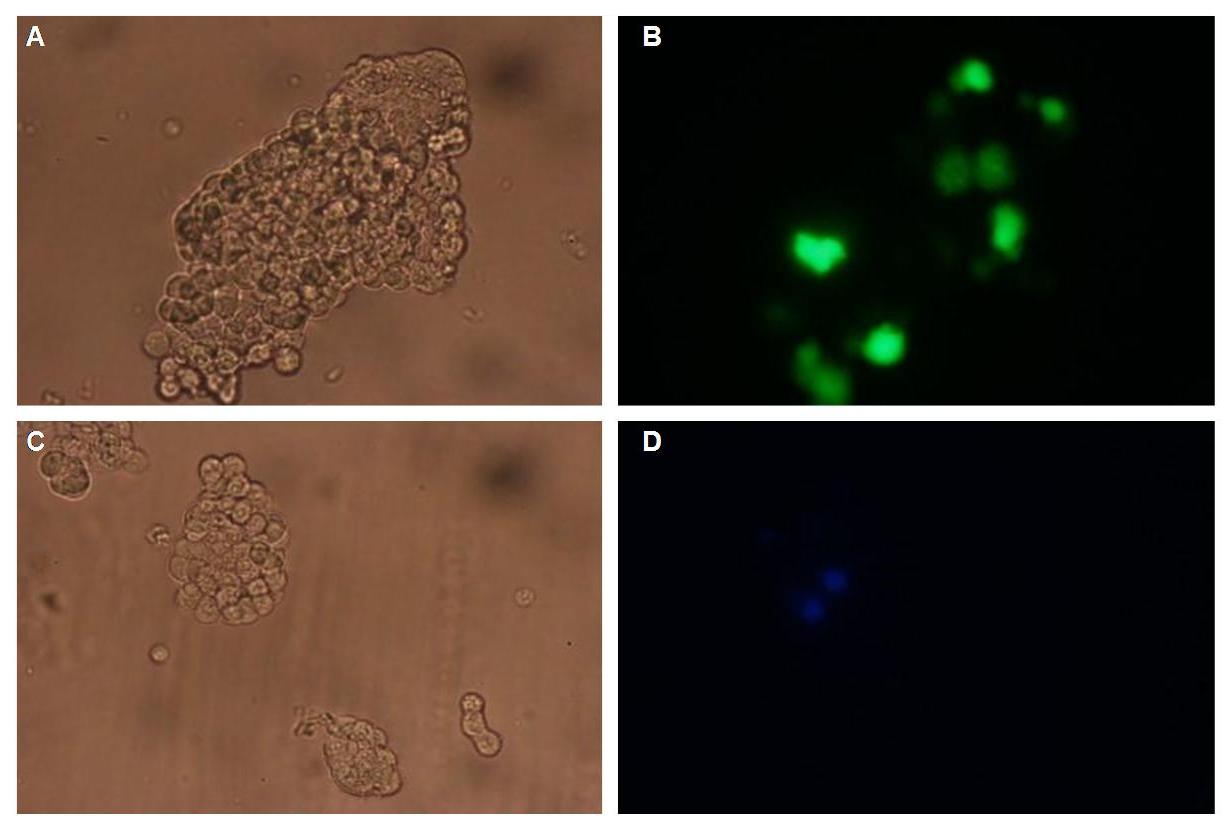
Figure_fluorescence microscopy 2 brightfield and UV-light picture with YFP- and CFP-filter. The 293T cells are transfected with the constructs: upper:Transfectionsvector+CMV_YFP lower:Transfectionsvector+CMV_CFP
The pictures show cleary the difference between soluble YFP/CFP Figure_fluorescence microscopy 2 and the membranelocalised YFP fused to the synthetic receptor construct Figure_fluorescence microscopy 1 "transfectionsvector+CMV_SP_Lipocalin_GGGS-linker_transmembraneregion_beta-Lactamase-fragment1_YFP". The principle to localise our synthetic fusion-receptor-proteins to cell membrane is proved.
|
 "
"
























