Team:Heidelberg/Notebook/Killing II/11thweek
From 2008.igem.org
(Difference between revisions)
(→Colicin activity test) |
(→Sunday 10/19/2008) |
||
| (8 intermediate revisions not shown) | |||
| Line 509: | Line 509: | ||
**Amplifier OD: <br> [[Image:081013-amplifier_od_small.jpg |500 px | center]] | **Amplifier OD: <br> [[Image:081013-amplifier_od_small.jpg |500 px | center]] | ||
**In most cases the later supernatants have a higher GFP/OD rate: But there in some cases earlier concentrations are much higher. A problem was that the plate stand at 4 °C for 2 days so this could cause these outliers. In the next days we repeat this experiment to characterize these parts properly. | **In most cases the later supernatants have a higher GFP/OD rate: But there in some cases earlier concentrations are much higher. A problem was that the plate stand at 4 °C for 2 days so this could cause these outliers. In the next days we repeat this experiment to characterize these parts properly. | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/11thweek back]] | ||
==Tuesday 10/14/2008== | ==Tuesday 10/14/2008== | ||
| Line 598: | Line 600: | ||
===Sender cloning: constitutive promotor-sender=== | ===Sender cloning: constitutive promotor-sender=== | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/11thweek back]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Wednesday 10/15/2008== | ==Wednesday 10/15/2008== | ||
| Line 639: | Line 633: | ||
pBAD-mCherry + T9002_without_GFP-Receiver in MG1655 | pBAD-mCherry + T9002_without_GFP-Receiver in MG1655 | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/11thweek back]] | |
| - | + | ||
==Thursday 10/16/2008== | ==Thursday 10/16/2008== | ||
| Line 741: | Line 734: | ||
====Colicin activity test==== | ====Colicin activity test==== | ||
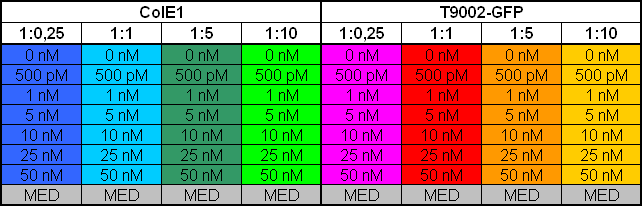
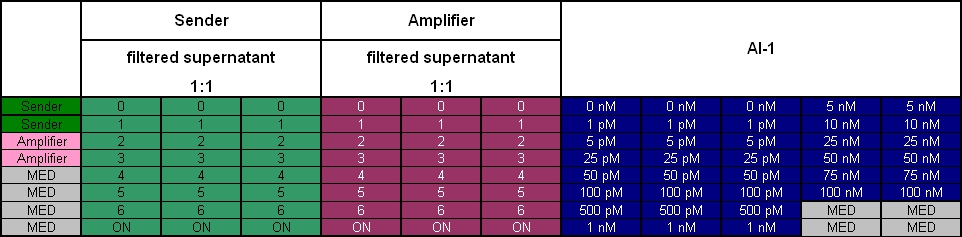
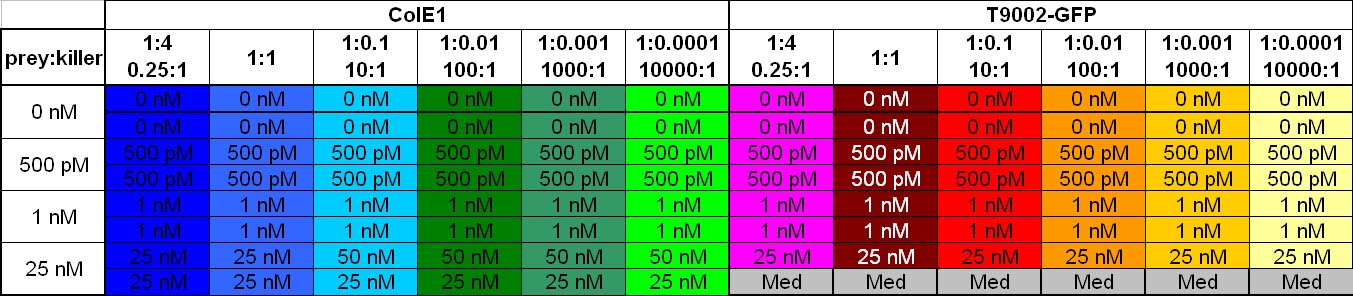
| + | After adjustment of the OD of the different cultures (killer, reference killer, prey), mixtures of different rations were done and transfered to a 96 well plate for ON measurement after following schemes | ||
| - | [[Image: 081016_colE1_test_legend.jpg | | + | [[Image: 081016_colE1_test_legend.jpg | 680 px | center]] |
| - | [[Image: 081016_colE1_test.jpg | | + | [[Image: 081016_colE1_test.jpg | 800 px | center]] |
===Sender=== | ===Sender=== | ||
*Results of sender test | *Results of sender test | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/11thweek back]] | |
| - | + | ||
| - | + | ||
==Friday 10/17/2008== | ==Friday 10/17/2008== | ||
| Line 765: | Line 757: | ||
===Sender Cloning: constitutive promotor - sender=== | ===Sender Cloning: constitutive promotor - sender=== | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/11thweek back]] | ||
==Saturday 10/18/2008== | ==Saturday 10/18/2008== | ||
| Line 837: | Line 830: | ||
====Sender and Amplifier Activity Test - 24h==== | ====Sender and Amplifier Activity Test - 24h==== | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/11thweek back]] | |
| - | + | ||
| - | + | ||
==Sunday 10/19/2008== | ==Sunday 10/19/2008== | ||
| Line 914: | Line 905: | ||
===Sender Cloning: constitutive promotor - sender=== | ===Sender Cloning: constitutive promotor - sender=== | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/11thweek back]] | ||
| + | |||
[[Team:Heidelberg/Notebook/Killing_II/12thweek | go to 12<sup>th</sup> week]] | [[Team:Heidelberg/Notebook/Killing_II/12thweek | go to 12<sup>th</sup> week]] | ||
Latest revision as of 21:50, 28 October 2008


11th week
Contents |
Monday 10/13/2008
Construction of reporterplasmid with kanamycin resistance
mCherry
- Transformationresults: Cultures were grown, also on plates with wrong plasmid - insert ratio
- Inoculation of liquid cultures from transformation with wrong ratio in the morning.
- Miniprep of liquid cultures in the evening: eluted in 35 µl H2O, (Qiagen Miniprepkit)
- Inoculation of liquid cultures from transformation with right ratio.
pSB1A2-Receiver-Colicin cloning
Mutagenesis of pSB1A2-ColicinE1-Receiver
- MiniPreps of colE1 after the 2nd mutagenesis (colonies 1.14, 1.16, 5.1, 5.7) and of colE9 (colonies 2 and 4, still with the pre-/suffix) each on pSB1A2 after receiver --> sent for sequencing
- Colonies picked from the transformations of the mutated colE9 parts (with the correct pre-/suffix) --> inocculated and MiniPreped (for digestion and sequencing)
Characterization: ColicinE1-Receiver Activitytest
- 4 pm: Inoculation of:
- 30 ml TB-Kana with BBa_I20260
- 30 ml TB-Amp with T9002-Receiver without GFP
- 30 ml TB-Amp with pSB1A2-ColicinE1-Receiver
- 8.30 pm: Preparing mixtures for the plate:
- plate scheme:
- Killer:Prey 1:0.25 -> 200 µl Referencepromotercells + 800 µl Receivercells
- Killer:Prey 1:1 -> 200 µl Referencepromotercells + 200 µl Receivercells + 600 µl TB media
- Killer:Prey 1:5 -> 200 µl Referencepromotercells + 40 µl Receivercells + 760 µl TB media
- Killer:Prey 1:10 -> 200 µl Referencepromotercells + 20 µl Receivercells + 780 µl TB media
- 9.30 pm: Starting measurement
Sender cloning: constitutive promotor-sender
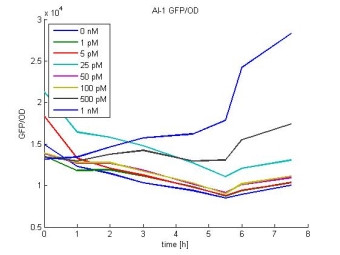
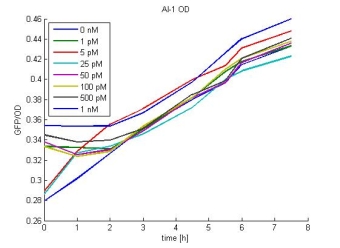
Characterization: Sender Activity Test
- Characterization: Sender-Test measured every hour over the day. See Saturdayurday for more details.
- Layout of the 96 well plate:
- Results:
- AI-1 GFP/OD:
- AI-1 OD:
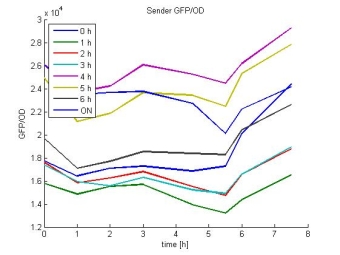
- Sender GFP/OD:
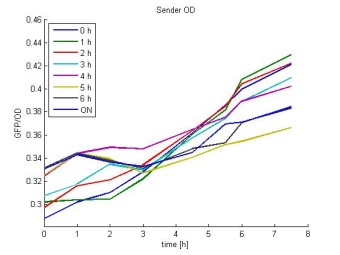
- Sender OD:
- Amplifier GFP/OD:
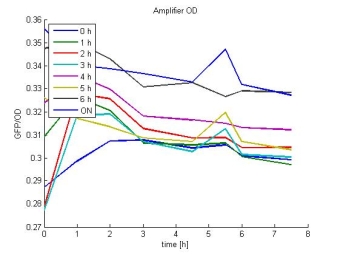
- Amplifier OD:
- In most cases the later supernatants have a higher GFP/OD rate: But there in some cases earlier concentrations are much higher. A problem was that the plate stand at 4 °C for 2 days so this could cause these outliers. In the next days we repeat this experiment to characterize these parts properly.
- AI-1 GFP/OD:
[back]
Tuesday 10/14/2008
Construction of reporterplasmid with kanamycin resistance
mCherry
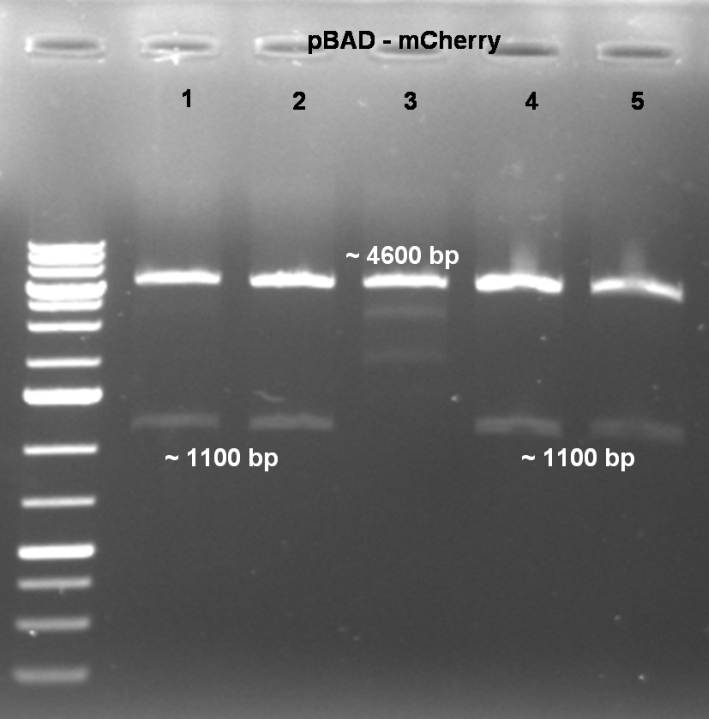
- Controldigestion of Cloning: 2 h 20 min -> 37 °C
10.0 µl pBAD-mCherry DNA (3rd mutation) 4.0 µl BSA 10x (NEB) 4.0 µl NEBuffer 4 (NEB) 0.5 µl SacI (NEB) 0.7 µl XbaI (NEB) 20.8 µl H2O ------- 40.0 µl
- Gel of digestion: 1% Agarose, 135 V, 30 min
- Results of Digestion:
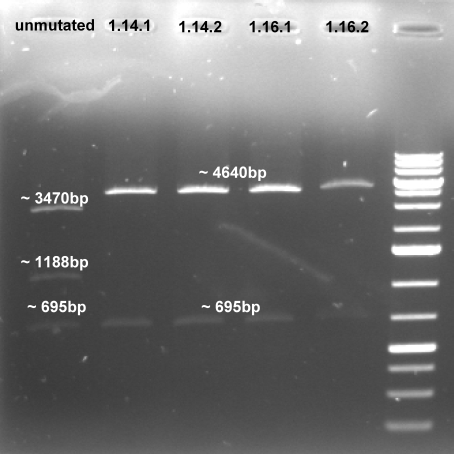
- Expected fragments for correct mutation: ~1100 bp & ~4600 bp
- Coolonies 1, 2, 4 & 5 shows the right bands. -> Sucessful cloning.
pSB1A2-Receiver-Colicin cloning
Mutagenesis of pSB1A2-Receiver-ColicinE1
- Miniprep of ON liquid cultures 3rd colicin E1 mutagenesis: eluted in 35 µl H2O, Qiagen Miniprepkit
- Controldigestion of 3rd Mutagenesis: 2 h 20 min -> 37 °C
10.0 µl ColE1-Receiver DNA (3rd mutation) 4.0 µl BSA 10x (NEB) 4.0 µl NEBuffer 3 (NEB) 0.5 µl PstI (NEB) 21.5 µl H2O ------- 40.0 µl
- Gel of digestion: 1% Agarose, 135 V, 30 min
- Results of Digestion:
- Expected fragments for correct mutation: ~695 bp & ~4600 bp
- The gel shows the right bands. This indicates that our three mutations were succesful.
- Mutagenesis PCR of Miniprep: Mutagenesis of 3rd PstI site
5.0 µl pfu buffer 1.0 µl forward primer (10 µM Col E1_mut_Pst_2_fw) 1.0 µl reverse primer (10 µM Col E1_mut_Pst_2_rv) 1.0 µl dNTPs (12.5 mM, 50x) 0.5 µl template DNA - 5 to 50 ng plasmid DNA 40.5 µl milliQ water 1.0 µl turbo pfu polymerase (Stratagene) ---- 50.0 µl
95 °C 30 sec 95 °C 30 sec | 55 °C 60 sec | 16 cycles 68 °C 11 min | 4 °C constant
- Digestion with DpnI (NEB) of the third mutagenesis to cut parental plasmid: Added 1 µl of DpnI to the finished Mutagenesis PCR. DpnI cuts at methylated GATC sites, so that only parental plasmids without the mutation are cutted. 2 h 35 min -> 37 °C
- Controlgel of third Mutagenesis (PstI_3): 1 % Agarose, 135 V, 30 min; DNA only visible in products from the colonie 1 (1.14 and 1.16) -> probably the mutagenesis worked on this plasmid.
- PCR Purification Kit (Qiagen) to purify the plasmids. Eluted in 35 µl H2O
- Transformation of prior mutagenesis: 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 500 µl on preheated LB-Amp plate - incubate overnight
Characterization: ColicinE1-Receiver Activitytest
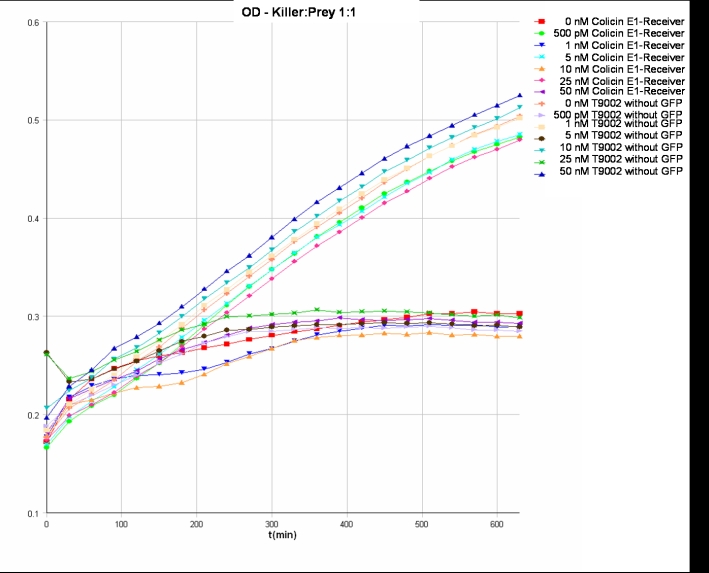
- Results:
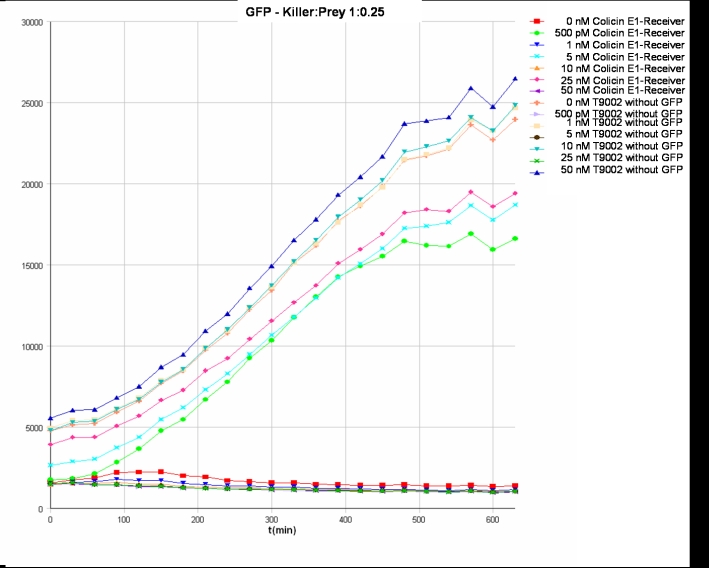
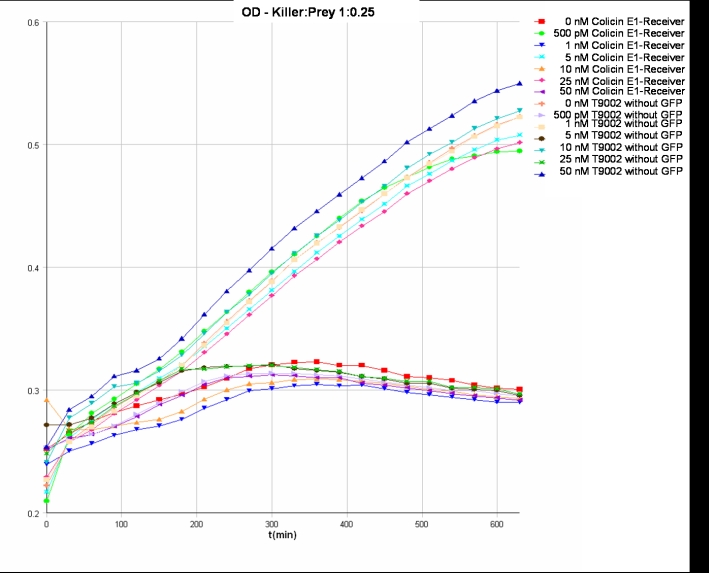
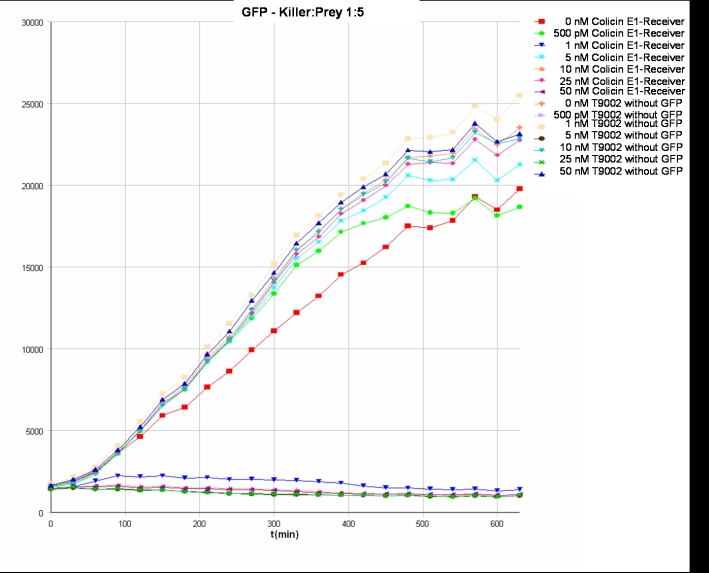
- Killer:Prey ratio 1:0.25 GFP:
- Killer:Prey ratio 1:0.25 OD:
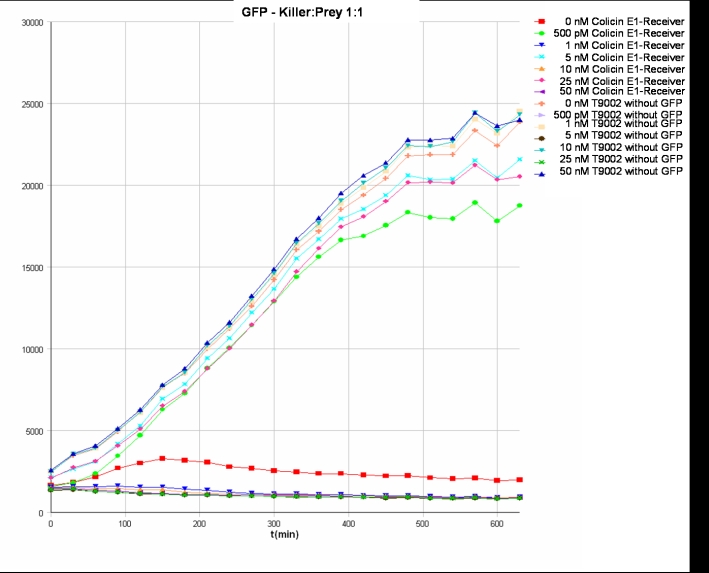
- Killer:Prey ratio 1:1 GFP:
- Killer:Prey ratio 1:1 OD:
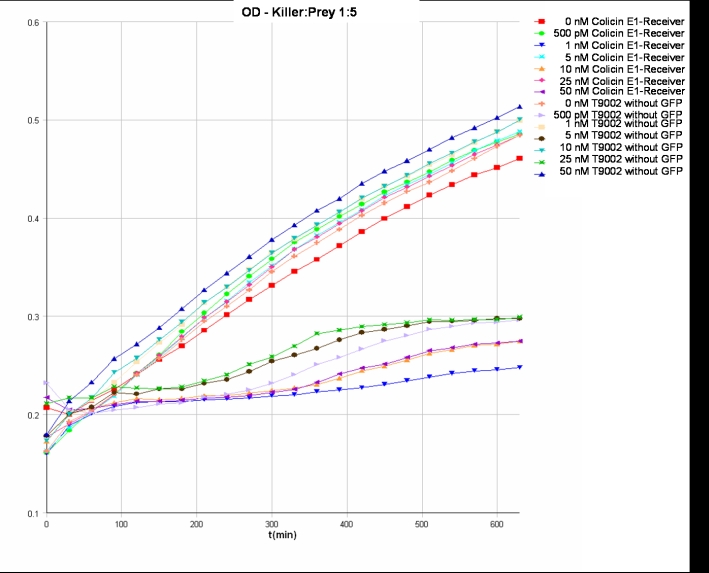
- Killer:Prey ratio 1:5 GFP:
- Killer:Prey ratio 1:5 OD:
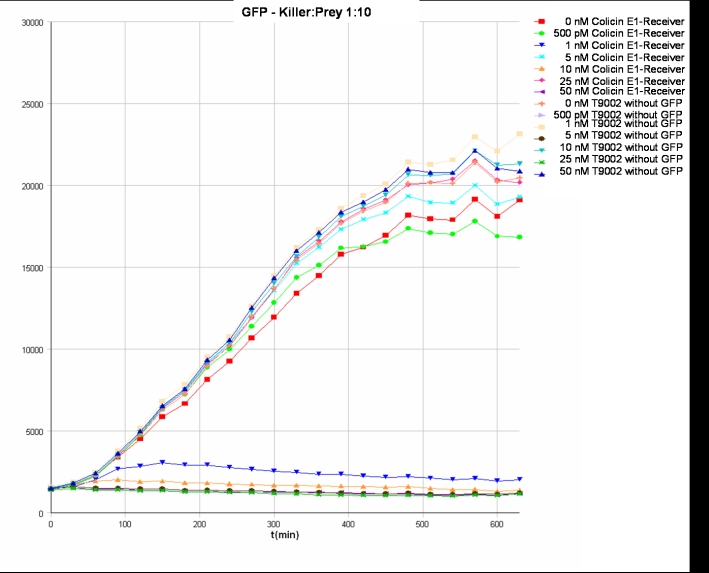
- Killer:Prey ratio 1:10 GFP:
- Killer:Prey ratio 1:10 OD:
- Killer:Prey ratio 1:0.25 GFP:
Sender cloning: constitutive promotor-sender
[back]
Wednesday 10/15/2008
Construction of reporterplasmid with kanamycin resistance
mCherry
- Sequencing results: Colonies 1, 2, 4 & 5 are positive clones. -> double transformation
pSB1A3-Receiver-Colicin cloning
Standardization
- Inoculation of 4th mutagenesis transformation cultures
- Miniprep of ColE9-Receiver BioBrickstandard: 5 colonies of ColE9-Receiver(2) and 5 colonies of ColE9-Receiver(4) (Qiagen, Miniprepkit) --> all 10 sent for sequencing
Activity Test
- Results
Sender
- Sendertest
Visualization
- Doubletransformation of:
I20260 + constitutive sender in TOP 10 I20260 in TOP 10 I20260 + ColicinE1-Receiver in TOP 10 I20260 + T9002_without_GFP-Receiver in TOP 10 pBAD-mCherry + ColicinE1-Receiver in TOP 10 pBAD-mCherry + T9002_without_GFP-Receiver in TOP 10 pBAD-mCherry + ColicinE1-Receiver in HCB33 pBAD-mCherry + T9002_without_GFP-Receiver in HCB33 pBAD-mCherry + ColicinE1-Receiver in MG1655 pBAD-mCherry + T9002_without_GFP-Receiver in MG1655
[back]
Thursday 10/16/2008
pSB1A2-Receiver-Colicin cloning
Sequencing results of ColE9-Receiver cloning
| EcoRI-site mutation | Prefix | Suffix | complete sequence | |
|---|---|---|---|---|
| colE9_(2) 1 | ? | ? | ? | ? |
| colE1_(2) 2 | + | + | + | not checked |
| colE1_(2) 3 | + | + | + | not checked |
| colE1_(2) 4 | + | + | + | not checked |
| colE1_(2) 5 | + | + | + | not checked |
| colE1_(4) 1 | + | + | + | + |
| colE1_(4) 2 | + | + | + | not checked |
| colE1_(4) 3 | ? | ? | ? | ? |
- Miniprep of ONC
- Controldigestion of Minipreps:
- Gel of Digestion:
- PCR of Minipreps:
2.0 µl DNA Template 2.5 µl Primer fw 2.5 µl Primer rv 18.0 µl H2O 25.0 µl Phusion MasterMix ------- 50.0 µl program:
98 °C 2 min 98 °C 10 sec | 53 °C 30 sec | 25 cycles 72 °C 90 sec | 72 °C 5 min 4 °C constant
- Gel of PCR
- Gelextraktion
- Ligation ColicinE1-Receiver (BioBrick Standard) with pSB1A2 (3:1): 1 h -> 22 °C
2.0 µl T4 DNA Ligase (Fermentas) 2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.6 µl pSB1A2 (6,5 ng/µl) 13.4 µl ColE1Receiver PCR Product (5,6 ng/µl)
- Transformation of prior ligation: 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 500 µl on preheated LB-Amp plate - incubate overnight
Colicin activity test
After adjustment of the OD of the different cultures (killer, reference killer, prey), mixtures of different rations were done and transfered to a 96 well plate for ON measurement after following schemes
Sender
- Results of sender test
[back]
Friday 10/17/2008
pSB1A3-Receiver-Colicin cloning
HisTag cloning of Colicins for purification
Sender Cloning: pBAD - sender
Sender Cloning: constitutive promotor - sender
[back]
Saturday 10/18/2008
pSB1A3-Receiver-Colicin cloning
ColE1 Cloning for standardization
- PCR-Amplification of Receiver-Col E1-Insert with the right Prefix and Suffix
25.0 µl Taq Master Mix (Fermentas) 2.5 µl T9002_LuxpR_Not_Eco_Xba_G_fw (Tm=80,96°C) 2.5 µl ColE1_kil_prot_rv_A_SpeI (Tm=66,94°C) 20.0 µl H2O 1 colony ------- 50.0 µl
program 1:
98 °C 10 sec 98 °C 30 sec | 57 °C 30 sec | 25 cycles 72 °C 1 min 45 sec | 72 °C 10 min 4 °C constant
program 2:
98 °C 10 sec 98 °C 30 sec | 67 °C 30 sec | 25 cycles 72 °C 1 min 45 sec | 72 °C 10 min 4 °C constant
- Gel of Receiver-colE1 PCR-amplification: 0,7% Agarose, 135 V, 30 min
- Gel-Results of colE1 PCR-amplification: Bands of PCR-A
- Gelextraction with Qiagen Kit: eluted in 40 µl H2O
- Digestion of receiver-colicinE1-insert for 2 h at 37°C
38.5 µl DNA (of Gelextraction) 0.5 µl EcoRI (20 000 U/ml, NEB) 1.0 µl SpeI (10 000 U/ml, NEB) 5.0 µl BSA 10x 5.0 µl EcoRI Buffer(NEB) ------- 50.0 µl
- PCR-Purification
- Ligation of receiver-colE1-insert into pSB1A2 (1):
1.Ligation attempt (+): 2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.0 µl T4 DNA Ligase (Fermentas) 1.8 µl pSB1A2 14.2 µl receiver-colE1-insert (From PCR-Purification) ------- 20.0 µl
2.Ligation attempt (-): 2.0 µl T4 DNA Ligase Buffer (Fermentas) 2.0 µl T4 DNA Ligase (Fermentas) 1.1 µl pSB1A2 14.9 µl receiver-colE1-insert (From PCR-Purification) ------- 20.0 µl
- Transformation of receiver-colE1-pSB1A2 BioBrick: 5 µl DNA (Ligation) per 50 µl E. coli TOP 10 competent cells
- start thawing 50 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 250 µl preheated LB media - incubate at 37 °C for 20 min, 500 rpm - Plate 300 µl on preheated LB-Amp plate - incubate overnight at 37°C
Sender Cloning: constitutive promotor - sender
Sender and Amplifier Activity Test - 24h
[back]
Sunday 10/19/2008
pSB1A3-Receiver-Colicin cloning
ColE1 Cloning for standardization
- Inoculation of colonies from transformation:
- 5 colonies from each plate in 10 ml LB-amp medium respectively:
- 57°C(+)-1
- 57°C(+)-2
- 57°C(-)-1
- 57°C(-)-2
- 67°C(+)-1
- 67°C(+)-3
- 67°C(-)-1
- 67°C(-)-3
- Minipreps:
- 57°C(+)-1
- 57°C(+)-2
- 57°C(-)-1
- 57°C(-)-2
- 67°C(+)-1
- 67°C(+)-3
- 67°C(-)-1
- 67°C(-)-3
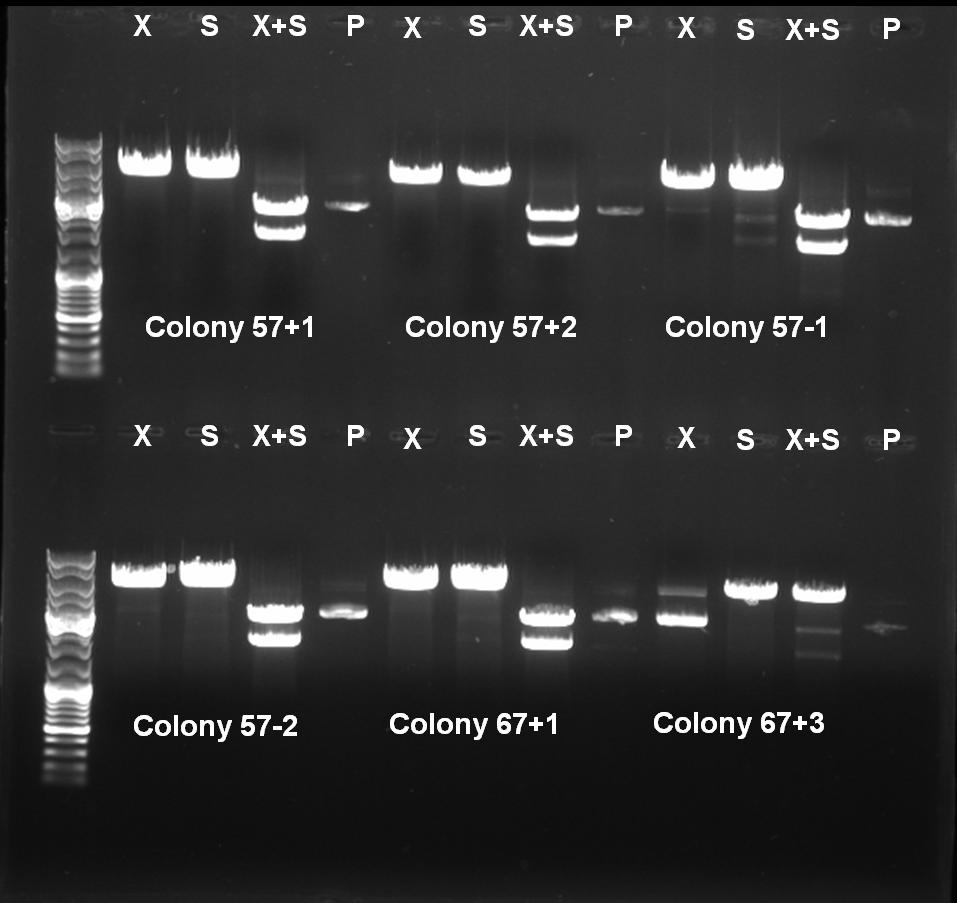
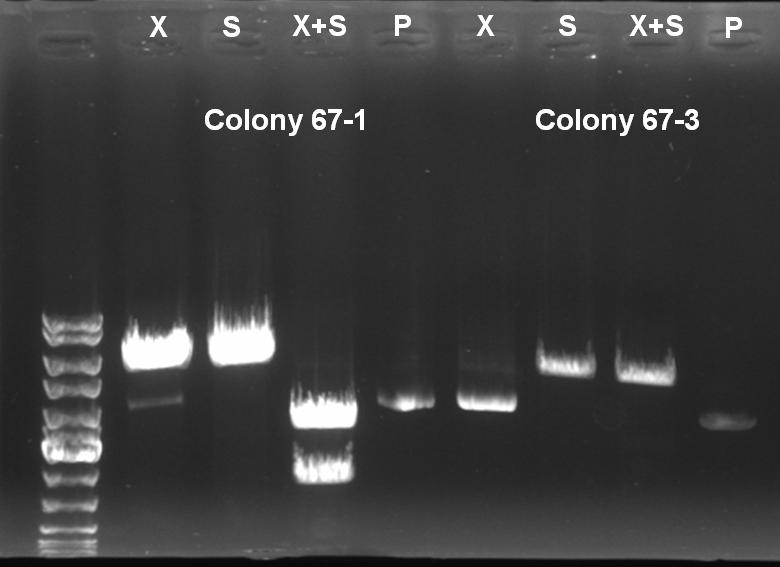
- Controldigestion with XbaI, SpeI and XbaI+SpeI of BioBrick Cloning: Receiver-ColicinE1-pSB1A3
XbaI digestion: 10.0 µl DNA 2.0 µl BSA 10x (NEB) 2.0 µl NEBuffer 2 0.5 µl XbaI (NEB) 5.5 µl H2O ------- 20.0 µl SpeI digestion: 10.0 µl DNA 2.0 µl BSA 10x (NEB) 2.0 µl NEBuffer 2 1.0 µl SpeI (NEB) 5.0 µl H2O ------- 20.0 µl SpeI digestion: 10.0 µl DNA 2.0 µl BSA 10x (NEB) 2.0 µl NEBuffer 2 0,5 µl XbaI 1.0 µl SpeI (NEB) 4.5 µl H2O ------- 20.0 µl
- Controlgel: 0,7% Agarose, 135 V, 30 min
- Gelresults:
- Expected Fragments:
- XbaI digestion (X): ~5200 bp (linear plasmid)
- SpeI digestion (S): ~5200 bp (linear plasmid)
- XbaI+SprI digestion (X+S): ~3200 bp (Insert) + ~2000 bp (Vector)
- Undigested Plasmid (P): ~3200 bp
- Expected Fragments:
- The following clones are positive ColE1 BioBricks:
- 57°C(+)-1
- 57°C(+)-2
- 57°C(-)-1
- 57°C(-)-2
- 67°C(+)-1
- 67°C(-)-1
- The following clones are positive ColE1 BioBricks:
Colicin Activity Test
Sender Cloning: constitutive promotor - sender
[back]
 "
"