Team:Imperial College/Biomaterial Details
From 2008.igem.org
m (Small grammar edits) |
|||
| (4 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
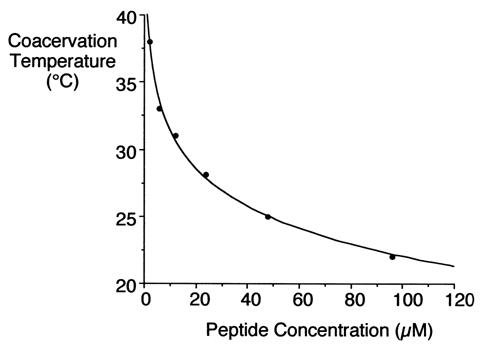
Under appropriate conditions of temperature and ionic strength, elastin undergoes a self-aggregation process called '''coacervation'''. This is where the protein separates from the solution as a second phase and is usually induced by an increase in temperature. | Under appropriate conditions of temperature and ionic strength, elastin undergoes a self-aggregation process called '''coacervation'''. This is where the protein separates from the solution as a second phase and is usually induced by an increase in temperature. | ||
| - | Unlike most proteins, which undergo denaturation when the temperature of the solution increases, elastin polypeptides become more ordered through coacervation. The temperature at which coacervation occurs is dependent on the relative proportions of hydrophobic and hydrophilic residues in the synthetic polypeptides as well as the ionic strength, pH and protein concentration of the solution. | + | Unlike most proteins, which undergo denaturation when the temperature of the solution increases, elastin polypeptides become more ordered through coacervation. The temperature at which coacervation occurs is dependent on the relative proportions of hydrophobic and hydrophilic residues in the synthetic polypeptides as well as the ionic strength, pH and protein concentration of the solution. ({{Ref|1}}, {{Ref|2}}) |
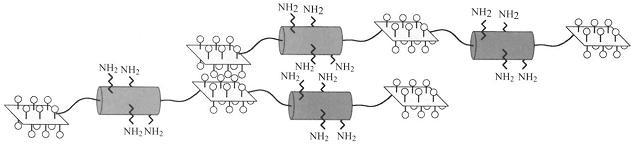
|[[Image:Elastin.JPG|300px|Construct EP20-24-24 for human elastin polypeptide]] | |[[Image:Elastin.JPG|300px|Construct EP20-24-24 for human elastin polypeptide]] | ||
| Line 18: | Line 18: | ||
{{Imperial/Box1|EAK16-II| | {{Imperial/Box1|EAK16-II| | ||
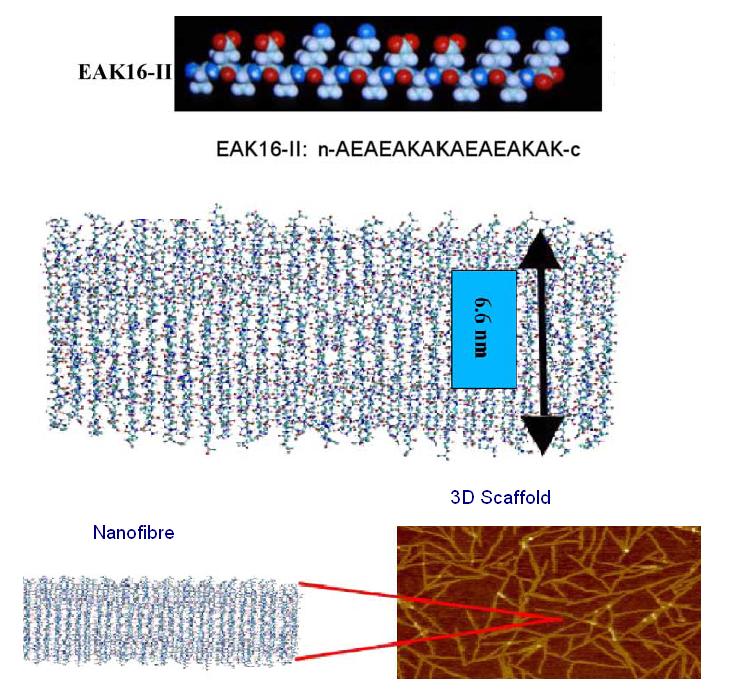
| - | EAK16-II is a self-assembling peptide which forms stable β-sheet structures in water and exists naturally as a region in zuotin, a yeast protein. The amino acid sequence for EAK16-II is AEAEAKAKAEAEAKAK and has an estimated molecular mass of 1615.8. When examined under SEM, a well-ordered nanofibre structure is formed by the association of the EAK16-II proteins and these nanofibres can | + | EAK16-II is a self-assembling peptide which forms stable β-sheet structures in water and exists naturally as a region in zuotin, a yeast protein. The amino acid sequence for EAK16-II is AEAEAKAKAEAEAKAK and has an estimated molecular mass of 1615.8. When examined under SEM, a well-ordered nanofibre structure is formed by the association of the EAK16-II proteins and these nanofibres can further aggregate to form a membranous 3D scaffold. |
| - | The alternating positive and negative charges (--++--++) are responsible for creating an electrostatic attraction between adjacent peptides and self-assembly is triggered when the proteins are exposed to physiological media or salt solution. | + | The alternating positive and negative charges (--++--++) are responsible for creating an electrostatic attraction between adjacent peptides and self-assembly is triggered when the proteins are exposed to physiological media or salt solution. ({{Ref|3}}) |
|[[Image:eak16.JPG|300px|Molecular structure of EAK16-II peptide]]}} | |[[Image:eak16.JPG|300px|Molecular structure of EAK16-II peptide]]}} | ||
{{Imperial/Box1|How ''B. subtilis'' can be used in producing material| | {{Imperial/Box1|How ''B. subtilis'' can be used in producing material| | ||
| - | B. subtilis can act as a cell factory producing and secreting proteins. Proteins can be secreted via three major pathways within | + | B. subtilis can act as a cell factory producing and secreting proteins. Proteins can be secreted via three major pathways within B. subtilis: Secretory signal recognition particle ('''Sec-SRP''') pathway, Twin-arginine translocation ('''Tat''') pathway and the ATP-binding cassette ('''ABC''') transporters. The Sec-SRP pathway would be more suitable in this case because it has been well studied in comparison to the two latter pathways. |
| - | ====== The process of Sec-SRP-dependent | + | ====== The process of Sec-SRP-dependent protein secretion ====== |
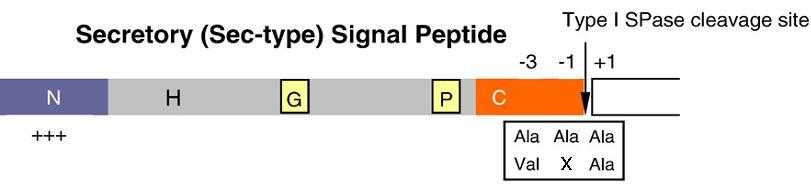
[[Image:signalpeptide.JPG|right|300px|Schematic representation of signal peptides for the Sec-SRP pathway in B. subtilis]] | [[Image:signalpeptide.JPG|right|300px|Schematic representation of signal peptides for the Sec-SRP pathway in B. subtilis]] | ||
# SRP-protein targeting system to cell membrane | # SRP-protein targeting system to cell membrane | ||
| Line 34: | Line 34: | ||
====== Signal Peptides ====== | ====== Signal Peptides ====== | ||
| - | Signal peptides are one of the major players in the production process of proteins. They are responsible for directing preproteins, a secretory protein with a signal peptide region attached, into a pathway. | + | Signal peptides are one of the major players in the production process of proteins. They are responsible for directing preproteins, a secretory protein with a signal peptide region attached, into a pathway. It functions to stop nascent chains mal-folding, recognising and submitting the peptide chain to the secretory machinery, and to act as the topological determinant for preproteins in the membrane. In the case of the sec-type signal peptide, they direct pre-proteins from the cytoplasm into the growth medium. Although different signal peptides show little similarity in amino acid sequence, three distinct domains can be distinguished: a positively charged N-terminus (N-region), a central hydrophobic region (H-region) and a polar C-terminal region (C-region). The C-region commonly carries a type-I SPase cleavage site, with the consensus sequence Ala-X-Ala or Val-X-Ala at positions -1 and -3 relative to the cleavage site. |
| - | At present, about 60% of all commercially available enzymes are produced by Bacillus bacteria due to their huge capacity for secretion. Heterologous proteins that were successfully secreted by Bacillus bacteria include cutinase, α-amylase, proinsulin | + | At present, about 60% of all commercially available enzymes are produced by Bacillus bacteria due to their huge capacity for secretion. Heterologous proteins that were successfully secreted by Bacillus bacteria include cutinase, α-amylase, and proinsulin ({{Ref|4}}, {{Ref|5}}) |
We have shortlisted the following three signal peptides to be transcribed upstream of our gene product: | We have shortlisted the following three signal peptides to be transcribed upstream of our gene product: | ||
| Line 48: | Line 48: | ||
<br><br> | <br><br> | ||
Go back to the <html><a href="https://2008.igem.org/Team:Imperial_College/Chassis_1"></html>'''>>> Specifications Page >>>'''<html></a></html>|}} | Go back to the <html><a href="https://2008.igem.org/Team:Imperial_College/Chassis_1"></html>'''>>> Specifications Page >>>'''<html></a></html>|}} | ||
| + | |||
| + | <hr><br clear="all"> | ||
| + | {{Imperial/Box2|| | ||
| + | =====References===== | ||
| + | # {{FormatRef|Bellingham CM, Woodhouse KA, Robson P, Rothstein SJ & Keeley FW|2001|Self-aggregation characteristics of recombinantly expressed human elastin polypeptides|Biochimica et Biophysica Acta 1550(1)|6-19||}} ([http://www.ncbi.nlm.nih.gov/pubmed/11738083 PubMed]) | ||
| + | # {{FormatRef|Keeley FW, Bellingham CM, & Woodhouse KA|2002|Elastin as a self-organizing biomaterial: use of recombinantly expressed human elastin polypeptides as a model for investigations of structure and self-assembly of elastin|Philos Trans R Soc Lond B Biol Sci. 357(1418)|185-189||}} ([http://www.ncbi.nlm.nih.gov/pubmed/11911775 PubMed]) | ||
| + | # {{FormatRef|Zhang S, Gelain F, & Zhao X|2005|Designer self-assembling peptide nanofiber scaffolds for 3D tissue cell cultures.|Semin Cancer Biol. 15(5)|413-420||}} ([http://www.ncbi.nlm.nih.gov/pubmed/16061392 PubMed]) | ||
| + | # {{FormatRef|Ling Lin Fu, Zi Rong Xu, Wei Fen Li, Jiang Bing Shuai, Ping Lu & Chun Xia Hu|2007|Protein secretion pathways in Bacillus subtilis: implication for optimization of heterologous protein secretion|Biotechnol Adv. 25(1)|1-12||}} ([http://www.ncbi.nlm.nih.gov/pubmed/16997527 PubMed]) | ||
| + | # {{FormatRef|Tjalsma H, Bolhuis A, Jongbloed JD, Bron S & van Dijl JM|2000|Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome|MicrobiolM Mol Biol Review 64(3)|515-547||}} ([http://www.ncbi.nlm.nih.gov/pubmed/10974125 PubMed]) | ||
| + | |}} | ||
| + | |||
| + | {{Imperial/EndPage|Chassis_1|Chassis_1}} | ||
Latest revision as of 03:46, 27 July 2009
|
|||||||||||||||||||||||
 "
"