Team:Davidson-Missouri Western/DNA Encoded XOR Gates
From 2008.igem.org
(New page: '''Biological Hash Function Designs''' - Our team conceived of five different ways to build genetic circuits that function as biological XOR gates. These could be used to produce a biolog...) |
Macampbell (Talk | contribs) |
||
| (2 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western|Home]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Team|The Team]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/New_Parts_Contributed_to_the_Registry|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Notebook|Notebook]] | ||
| + | |} | ||
| + | <br> | ||
'''Biological Hash Function Designs''' - Our team conceived of five different ways to build genetic circuits that function as biological XOR gates. These could be used to produce a biological hash function. | '''Biological Hash Function Designs''' - Our team conceived of five different ways to build genetic circuits that function as biological XOR gates. These could be used to produce a biological hash function. | ||
| Line 24: | Line 32: | ||
4. presence/absence of degradation tag on proteins | 4. presence/absence of degradation tag on proteins | ||
| - | [[Image:XOR DR AI2.PNG]] | + | <center>[[Image:XOR DR AI2.PNG]]</center> |
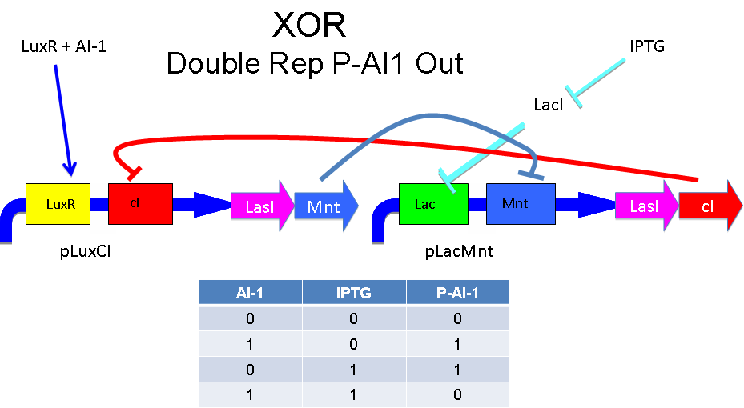
'''Above '''- Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed. | '''Above '''- Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed. | ||
| - | + | <center> | |
[[Image:XOR DR AI1b.PNG]]<br> | [[Image:XOR DR AI1b.PNG]]<br> | ||
| + | </center> | ||
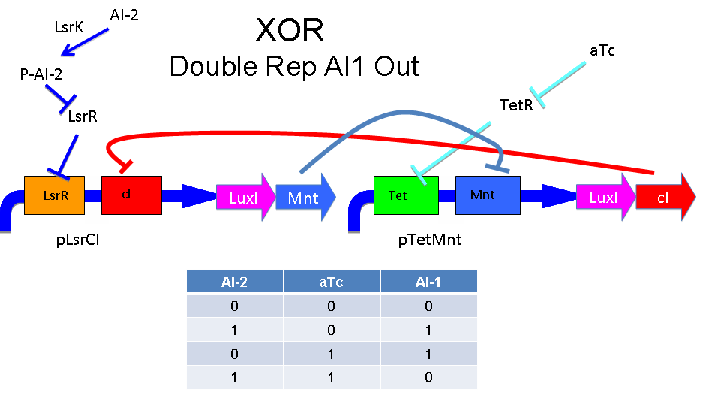
'''Above''' - Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed. | '''Above''' - Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed. | ||
| Line 39: | Line 48: | ||
| - | + | <center> | |
[[Image:xors dr pai1.PNG]] | [[Image:xors dr pai1.PNG]] | ||
| - | + | </center> | |
'''Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed. | '''Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed. | ||
| Line 47: | Line 56: | ||
| - | + | <center> | |
[[Image:xors dr ai1.PNG]] | [[Image:xors dr ai1.PNG]] | ||
| - | + | </center> | |
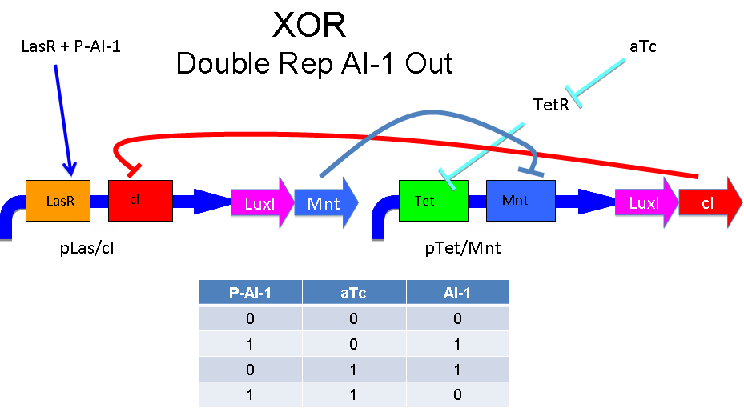
'''Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed.''' | '''Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed.''' | ||
| + | <br> | ||
| + | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western|Home]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Team|The Team]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/New_Parts_Contributed_to_the_Registry|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Notebook|Notebook]] | ||
| + | |} | ||
Latest revision as of 01:41, 30 October 2008
| Home | The Team | E. nigma Project | Parts Submitted to the Registry | Notebook |
|---|
Biological Hash Function Designs - Our team conceived of five different ways to build genetic circuits that function as biological XOR gates. These could be used to produce a biological hash function.
LuxR XOR Biological Design
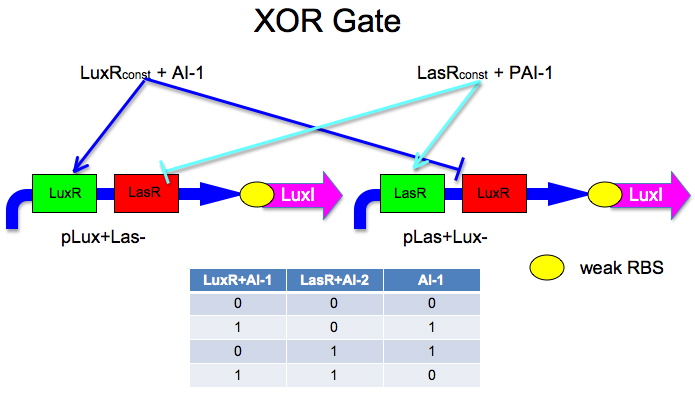
The idea is to have two mirrored halves of the system. LasR is regulated by PAI-1 {3-oxododecanoyl-HSL (3OC12HSL)} and LuxR is activated by AI-1 {3-oxohexanoyl-homoserine lactone ([http://partsregistry.org/3OC6HSL 3OC6HSL])}. There is a potential problem in that the Lux half is more likely to get positive feedback than the Las half. This MAY not be a problem because 0/0 is leaky so we put a weak RBS to minimize leaky protein production. Also, if we add AI-2 and AI-1 is produced by leak, then the entire system shuts down. The repressor site is located between -35 and -10 of the promoter. The activator binding site is upstream of -35. This has been documented [http://www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf by Egland and Greenberg]
Lsr/Lux XOR Biological Design
These two XOR circuits are designed to complement each other. Each recieves a cell-to-cell signal (AI-1 or AI-2) and a chemical signal (IPTG or AHL) and processes it into a cell-to-cell signal. Colonies that output AI-1 would alternate with colonies that produce AI-2. The input message to be hashed could be encoded by the presence or absence of the chemical signals, which would also alternate.
Design Variables
1. strength on RBS for each of the coding sequences (eg. RBS for enzymes could be weak)
2. order of coding sequences (eg. enzymes could be second for lower expression level)
3. identity of repressors (eg. mutant LacI repressors)
4. presence/absence of degradation tag on proteins
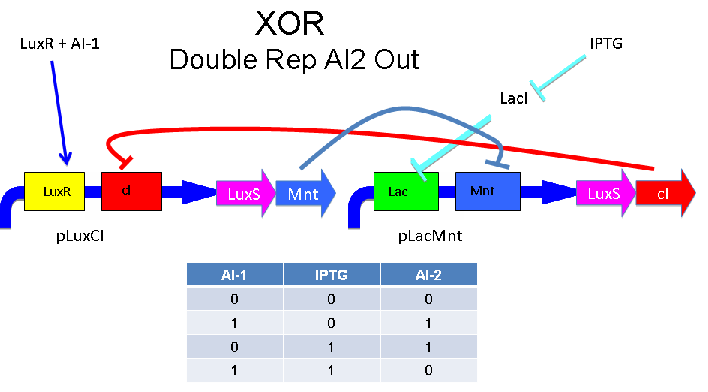
Above - Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed.
Above - Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed.
Las/Lux XOR Biological Design
A strength of the Davidson XOR design above is that each of the two signalling molecules used is foreign to E. coli. A strength of the Missouri Western design above is that is uses two complementary XOR gates. The design below combines the two approaches. Each XOR circuit receives a cell-to-cell signal (AI-1 or P-AI-1) and a chemical signal (IPTG or AHL) and processes it into a cell-to-cell signal. Colonies that output AI-1 would alternate with colonies that produce P-AI-1. The input message to be hashed could be encoded by the presence or absence of the chemical signals, which would also alternate.
Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed.
Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed.
| Home | The Team | E. nigma Project | Parts Submitted to the Registry | Notebook |
|---|
 "
"