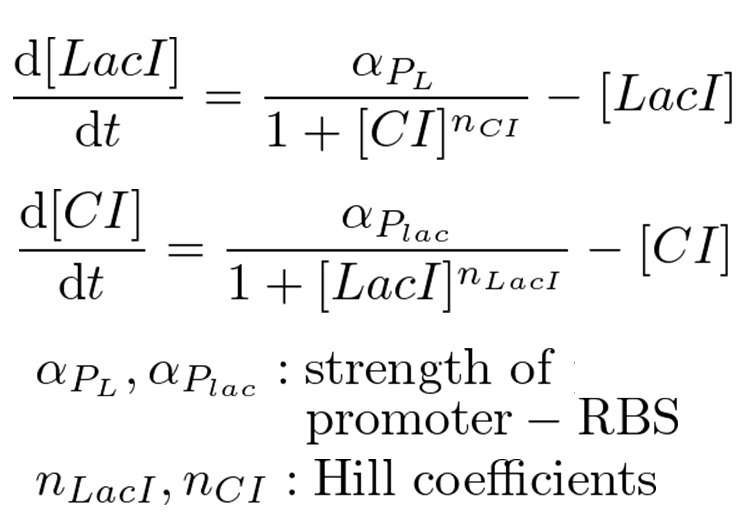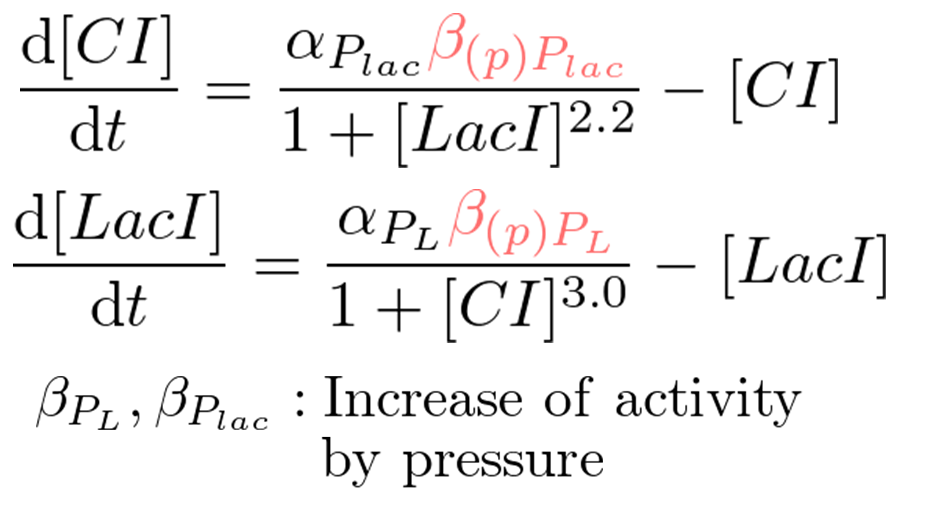Team:Tokyo Tech
From 2008.igem.org
(→Why did we use mathematical model ?) |
(→3. Touch display) |
||
| (252 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="90%" align="center" | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="90%" align="center" | ||
!align="center" width=20% style="text-align:center"|[[Team:Tokyo_Tech|Main]] | !align="center" width=20% style="text-align:center"|[[Team:Tokyo_Tech|Main]] | ||
| + | !align="center" width=20% style="text-align:center"|[[Team:Tokyo_Tech/Protocol|Protcol]] | ||
!align="center" width=20% style="text-align:center"|[[Team:Tokyo_Tech/Parts|Parts Submitted to the Registry]] | !align="center" width=20% style="text-align:center"|[[Team:Tokyo_Tech/Parts|Parts Submitted to the Registry]] | ||
!align="center" width=20% style="text-align:center"|[[Team:Tokyo_Tech/Team|Our Team]] | !align="center" width=20% style="text-align:center"|[[Team:Tokyo_Tech/Team|Our Team]] | ||
| Line 11: | Line 12: | ||
| - | == <font size=5>'''Our project''' </font>== | + | |
| + | == <font size=5>'''1. Our project''' </font>== | ||
<html> | <html> | ||
<body> | <body> | ||
| + | |||
| + | <table width="100%" border="0" cellspacing="0" cellpadding="0" > | ||
| + | |||
| + | <tr> | ||
| + | |||
| + | <td class="a" width="2%"> | ||
| + | </td> | ||
| + | <td class="a"> | ||
| + | <div> | ||
| + | <b> | ||
| + | <font size=4>Our project is creation of "<i style='mso-bidi-font-style:normal'>Coli</i> Touch"!!</font> | ||
| + | </b> | ||
| + | </div> | ||
| + | </td> | ||
| + | |||
| + | <td> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/f/f8/Tech_figure2.jpg" width="400" align="right"> | ||
| + | </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | </table> | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0" > | <table width="100%" border="0" cellspacing="0" cellpadding="0" > | ||
| - | + | <tr> | |
| - | + | ||
| - | + | <td> | |
| - | < | + | <img src="https://static.igem.org/mediawiki/2008/5/57/Tech_Coli_Touch.jpg" width="300" align="left"> |
</td> | </td> | ||
| - | |||
| - | |||
| - | + | <td class="a" width="2%"> | |
| - | + | </td> | |
| + | <td class="a"> | ||
| + | |||
| + | <p><div><b>What is "<i style='mso-bidi-font-style:normal'>Coli</i> touch"?</b></div></p> | ||
| + | <div>“<i style='mso-bidi-font-style:normal'>Coli</i> Touch” has a pressure sensitive display composed of an <i style='mso-bidi-font-style:normal'>E. coli</i> lawn. When you touch its display, touched section is colored.</div> | ||
| + | <div>Next I'll tell you about “<i style='mso-bidi-font-style:normal'>Coli</i> Touch” work system. Display of “<i style='mso-bidi-font-style:normal'>Coli</i> Touch” contains many <i style='mso-bidi-font-style:normal'>E. coli</i>. When you touch this display, pressure applies to <i style='mso-bidi-font-style:normal'>E. coli</i> in this display. Pressure applied <i style='mso-bidi-font-style:normal'>E. coli</i> expresses GFP. | ||
| + | </div><p><div><b>Why pressure?</b></div></p> | ||
| + | <p><div>“<i style='mso-bidi-font-style:normal'>Coli</i> Touch” uses pressure as input. Why do we use pressure? Past input methods (small molecules, heat and light) are difficult to induce uniformly. | ||
| + | Pressure can induce uniformly. | ||
| + | </div></p> | ||
| + | |||
| + | </td> | ||
| + | |||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | == <font size=5>'''2. Pressure induction'''</font> == | ||
| + | |||
| + | <font size=3>'''Introduction'''</font> | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
<td class="a" width="2%"> </td> | <td class="a" width="2%"> </td> | ||
<td class="a"> | <td class="a"> | ||
| - | + | <div>Tet promoters is known as a sensitive ones to pressure. (T. Sato et al., 1995)</div><td> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</tr> | </tr> | ||
| - | + | </table> | |
| - | + | ||
</body> | </body> | ||
</html> | </html> | ||
| - | |||
| - | |||
<font size=3>'''Construction'''</font> | <font size=3>'''Construction'''</font> | ||
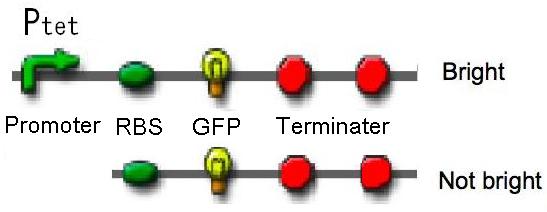
| - | [[Image: | + | [[Image:Tech_Tet_figure1.jpg|400px|thumb|right|figure2-1. We constructed Ptet-GFP and promoter less-GFP.]] |
<html> | <html> | ||
<body> | <body> | ||
| Line 66: | Line 97: | ||
<td class="a" width="3%"> </td> | <td class="a" width="3%"> </td> | ||
<td class="a"> | <td class="a"> | ||
| - | <div> | + | <div> We chose the tet promoter (Ptet) as a pressure-inducible promoter. We constructed two plasmids - one is Ptet-GFP on <a href="http://partsregistry.org/Part:BBa_I739201">pSB6</a>. The other is promoter less-GFP on <a href="http://partsregistry.org/Part:BBa_I739201">pSB6</a> as a negative control. We measured activity of tet promoters under 0.1 MPa and 30 MPa without repressor protein.</div></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 80: | Line 111: | ||
{{clear}} | {{clear}} | ||
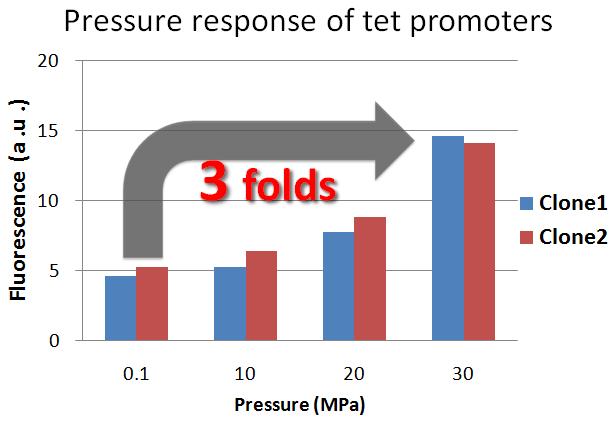
| - | <font size=3>'''Result | + | <font size=3>'''Result -activity of tet promoters- '''</font> |
| - | [[Image: | + | |
| + | [[Image:Tech Pressure response of tet promoter1.jpg|450px|thumb|right|figure2-2. Pressure response of the tet promoter without repressor protein. Tet promoters' activity increased under 30 MPa]] | ||
<html> | <html> | ||
<body> | <body> | ||
| Line 88: | Line 120: | ||
<td class="a" width="2%"> </td> | <td class="a" width="2%"> </td> | ||
<td class="a"> | <td class="a"> | ||
| - | <div>The result shows that | + | <div>The result shows that the tet promoter activity under 30 MPa, without repressor protein, is about 3-fold stronger than the tet promoter activity under 0.1 MPa. Therefore, we confirmed that the tet promoter was induced under 30 MPa.</div><td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 109: | Line 141: | ||
<p> </p> | <p> </p> | ||
| - | == <font size=5>'''Touch display'''</font> == | + | == <font size=5>'''3. Touch display'''</font> == |
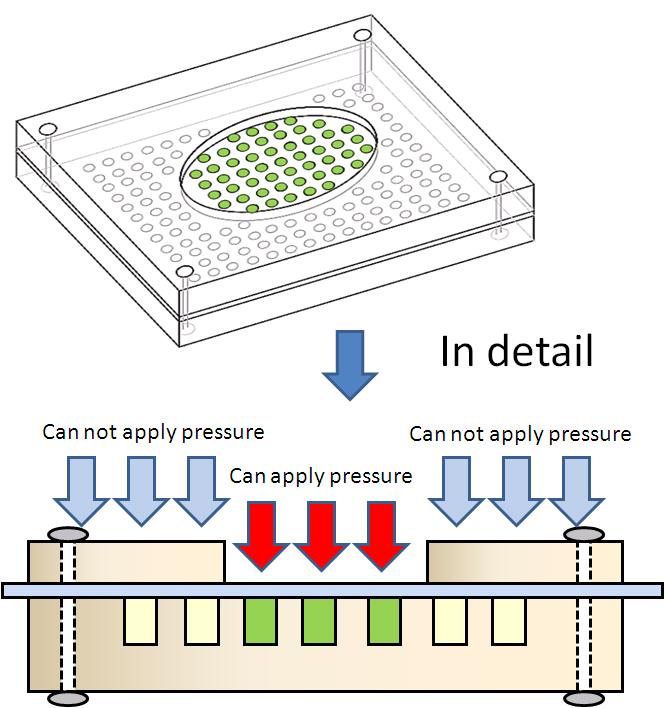
| - | <font size=3>''' | + | <font size=3>'''Touch display''' (plan)</font> |
| + | |||
| + | [[image:Tech Pressure device2.jpg|right|400px|thumb|figure 3-1-b. Design of touch display]] | ||
| + | |||
<html> | <html> | ||
| - | + | <body> | |
| - | + | <table width="55%" border="0" cellspacing="0" cellpadding="0"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <table width=" | + | |
<tr> | <tr> | ||
<td class="a" width="2%"> </td> | <td class="a" width="2%"> </td> | ||
<td class="a"> | <td class="a"> | ||
| - | < | + | <div>Touch display which we planned has many holes, and <i>E. coli</i> is in these holes.(figure 3-1 a)<div> |
| - | + | <div>As the first step of creating the touch display, we created Prototype touch display. figure 3-1 b)</div> | |
| + | </p></td> | ||
| + | |||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</body> | </body> | ||
</html> | </html> | ||
| + | <p> </p><p> </p> | ||
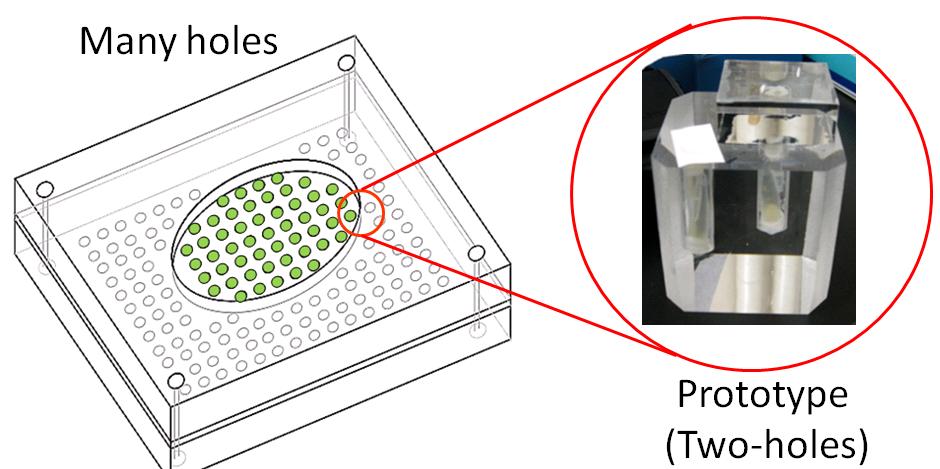
| - | <font size=3>''' | + | [[image:Tech Pressure device,jpg.jpg|left|400px|thumb|figure 3-1-a. Plan to create touch display. We created two-holes display as the first step.]] |
| + | |||
| + | |||
| + | {{clear}} | ||
| + | |||
| + | <font size=3>'''Prototype touch display'''</font> | ||
| + | |||
| + | [[image:Tech_aqulyl2.JPG|right|450px|thumb|figure 3-2. Prototype touch display. The display has two kinds of holes and two kinds of covers. ]] | ||
<html> | <html> | ||
<body> | <body> | ||
| - | <table width=" | + | <table width="45%" border="0" cellspacing="0" cellpadding="0"> |
| + | |||
<tr> | <tr> | ||
<td class="a" width="2%"> </td> | <td class="a" width="2%"> </td> | ||
<td class="a"> | <td class="a"> | ||
| - | < | + | <div class=MsoNormal>We created a Prototype touch display made of acrylic glasses. This touch display has two kinds of holes(show figure 3-2). Each hole contains culture medium and <i>E. coli</i> is cultivated in these holes. One hole (A) can be pressurized, because the hole is covered with only a plastic tape. Water pressure conducts into the hole.</div> |
| - | + | <div>The other (B) is not pressurized, because the hole is covered with a block made of acrylic glasses. Water pressure doesn’t conduct into the hole.</font></div> | |
| + | <td class="a" width="3%"> </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
| - | [[ | + | </body></html> |
| + | |||
| + | [[image:Tech_aqulyl1.jpg|left|thumb|800px|figure 3-3. Touch display. We pour culture medium into the holes.]] | ||
| + | <p> </p> | ||
| + | <p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p><p> </p> | ||
| + | <p> </p> | ||
| + | |||
| + | |||
| + | <font size=3>'''Result ~ <i>E. coli</i> in the touch display ~'''</font> | ||
| - | |||
<html> | <html> | ||
<body> | <body> | ||
| Line 169: | Line 202: | ||
<td class="a" width="2%"> </td> | <td class="a" width="2%"> </td> | ||
<td class="a"> | <td class="a"> | ||
| - | <p class=MsoNormal>After | + | <p class=MsoNormal>After incubation, we observed the <i>E. coli</i> by a fluorescence microscope.</p></td> |
<td class="a" width="3%"> </td> | <td class="a" width="3%"> </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </body></html> | ||
| - | + | | |
| - | + | ||
| - | + | [[image:Tech_apulyl_pic1.jpg|thumb|center|600px|figure 3-5. Images from fluorescence microscope. <i>E. coli</i> on left image is more bright than right one]] | |
| - | + | <html><body> | |
| - | + | ||
| - | < | + | |
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
| Line 185: | Line 217: | ||
<td class="a" width="2%"> </td> | <td class="a" width="2%"> </td> | ||
<td class="a"> | <td class="a"> | ||
| - | <div><b>The touch display successfully regulated GFP expression in E. coli | + | <div><b>The touch display successfully regulated GFP expression in <i>E. coli</i> !</b></div></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 196: | Line 228: | ||
</html> | </html> | ||
| - | == <font size=5>'''Low pressure-inducible promoter'''</font> == | + | == <font size=5>'''4. Low pressure-inducible promoter'''</font> == |
| - | [[Image: | + | [[Image:tokyotech2008Previous study - Pressure response of Plac.png|thumb|right|350px|box|Figure 4-1. - Pressure response of Plac]] |
<html> | <html> | ||
<body> | <body> | ||
<div> | <div> | ||
| - | It is known that lac promoter is induced under 30 MPa (T. Sato et al., 1995).However, 30 MPa is too high to | + | It is known that lac promoter is induced under 30 MPa (T. Sato et al., 1995). However, 30 MPa is too high to push with the fingers. |
</div> | </div> | ||
<div> | <div> | ||
| - | Therefore, we | + | Therefore, we tried to develop low pressure-inducible promoter. |
</div> | </div> | ||
</body> | </body> | ||
| Line 211: | Line 243: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | = | + | |
| - | <gallery widths="400px" heights=" | + | |
| - | Image: | + | |
| - | Image: | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <font size=3>'''Methods''' </font> | ||
| + | <gallery widths="400px" heights="230px" > | ||
| + | Image:Tokyotech2008PCR random mutagenesis.png|Figure 4-2 - PCR random mutagenesis | ||
| + | Image:Tokyotech2008Screening scheme with flow cytometer.png|Figure 4-3 - Screening scheme with flow cytometry | ||
</gallery> | </gallery> | ||
| - | We | + | We tried to develop a low pressure inducible promoter by PCR random mutagenesis to lac promoter. |
| - | + | Then we screened an ''E. coli'' library, with flow cytometry, for promoters that are induced under low pressure. | |
| - | This scheme is based on the ability to separate bacteria with a | + | This scheme is based on the ability to separate bacteria, with a flow cytometer, in response to expression, or lack of expression, of a fluorescent marker. |
| - | *''' | + | *'''Step 1''' - Fluorescent bacteria without repressor protein were collected by a flow cytometer. This sorted pool contains bacteria bearing both constitutive and low pressure-inducible promoter. |
| - | *''' | + | *'''Step 2''' - This sorted pool contains bacteria bearing both constitutive and pressure-inducible promoter fusions. False positives are removed with repressor protein. All non-fluorescent bacteria are sorted. |
| - | *''' | + | *'''Step 3''' - A final passage. We sorted bacteria under pressure with repressor protein. Fluorescent bacteria are sorted. False negatives are removed. Therefore bacteria bearing promoter that are low pressure-inducible are isolated. |
| - | = | + | |
| + | <font size=3>'''Results - Sequence and Characterization -''' </font> | ||
We finished step 1. | We finished step 1. | ||
Fluorescent and non-fluorescent bacteria were sorted and we characterized their promoter. | Fluorescent and non-fluorescent bacteria were sorted and we characterized their promoter. | ||
| - | + | ||
| - | <gallery widths="430px" heights=" | + | <gallery widths="430px" heights="240px" > |
| - | Image: | + | Image:Tokyotech2008Dot plot of cell sorting .png|figure 4-4. Dot plot of cell sorting |
| - | Image: | + | Image:Tokyotech2008Results of sequencing .png|figure 4-5. Results of sequencing |
</gallery> | </gallery> | ||
| - | We sorted fluorescent (A) and non-fluorescent bacteria (B) with a | + | We sorted fluorescent (A) and non-fluorescent bacteria (B) with a flow cytometer. |
Then, we analyze these base sequences. | Then, we analyze these base sequences. | ||
| - | *A have mutations in | + | *A have mutations in LacI binding site or non-functional DNA. |
*B have mutations in CAP binding site, -35 or non-functional DNA. | *B have mutations in CAP binding site, -35 or non-functional DNA. | ||
| + | Therefore, fluorescent bacteria have no mutation in CAP binding site, -35 or -10. | ||
| - | + | We have successfully demonstrated that it is possible to collect promoters desired functions by PCR random mutagenesis and screening with a flow cytometry. | |
| - | We have successfully demonstrated that it is possible to collect | + | This results indicate that we can screen low pressure-inducible lac promoter mutant with this methods. |
| - | + | ||
| - | == <font size=5>'''Write/Erase cycle'''</font> == | + | == <font size=5>'''5. Write/Erase cycle'''</font> == |
| - | [[Image:Write-Erase_cycle.png|thumb| | + | [[Image:Write-Erase_cycle.png|thumb|400px|right|figure 5-1. Write/Erase cycle]] |
| - | While we can implement write-function, we | + | While we can implement write-function, we want to implement additionally erase-function and memory-function. Erase-function enables us to erase the painted picture, and memory-function enables us to keep the picture after we stop induction. We call these functions "Write/Erase cycle". |
In order to implement Write/Erase cycle, we tried to construct genetic toggle switch. | In order to implement Write/Erase cycle, we tried to construct genetic toggle switch. | ||
{{clear}} | {{clear}} | ||
| - | |||
| - | |||
| - | + | === Genetic toggle switch to implement Write/Erase cycle === | |
| + | [[Image:Toggle_genetic_circuit_model.png|thumb|480px|right|figure 5-2. Genetic toggle switch]] | ||
| - | + | # '''Write-function''' | |
| + | ## '''P<sub>lac</sub>''' is under 30 MPa pressure. | ||
| + | ## P<sub>lac</sub> expresses CI and GFP. | ||
| + | ## CI represses P<sub>L</sub> and decreases LacI expression. | ||
| + | ## Low LacI expression increases P<sub>lac</sub> activity. ⇒ '''Bright!!''' | ||
| + | # '''Erase-function''' | ||
| + | ## The heat activates '''P<sub>L</sub>'''. | ||
| + | ## P<sub>L</sub> expresses LacI. | ||
| + | ## LacI represses Plac. | ||
| + | ## Therefore, GFP expression decreases. | ||
| - | + | {{clear}} | |
| - | + | === Mathematical model === | |
| + | ==== Why did we use mathematical model? ==== | ||
| + | [[Image:Conditions_for_Write-function.png|thumb|500px|right| figure 5-3. Left If P<sub>L</sub> is not activated or is a bit, write-function is available. Right If P<sub>L</sub> is activated too much, write-function is not available]] | ||
| - | + | As mentioned above, it is known that P<sub>lac</sub> is activated '''94'''-fold under 30 MPa while we didn't know the increase of P<sub>L</sub> strength under 30 MPa. If P<sub>L</sub> is activated too much and P<sub>lac</sub> activity is weaker than P<sub>L</sub> activity, we can't implement write-function. So, how much is the range of the increase of P<sub>L</sub> activity under 30 MPa so as to become advantageous to that of P<sub>lac</sub>? To know this range, we need to use mathematical model. | |
| - | + | {{clear}} | |
| - | + | ==== Classical toggle switch model ==== | |
| - | + | <gallery widths = "300px" heights="215px" perrow = "1" align = "right"> | |
| + | Image:Classical_toggle_switch_model.png| figure 5-4. Classical toggle switch model | ||
| + | Image:Transfer_Function.png| figure 5-5. Transfer function of P<sub>lac</sub> | ||
| + | </gallery> | ||
| - | + | Our mathematical model under 0.1 MPa is equal to a classical toggle switch model shown in figure 5-4. n<sub>CI</sub> is the cooperativity of repression of | |
| + | the lambda promoter, n<sub>LacI</sub> is the cooperativity of repression of the lac promoter, α<sub>PL</sub> is the effective rate of LacI synthesis | ||
| + | and α<sub>Plac</sub> is the effective rate of CI synthesis. | ||
| + | n<sub>CI</sub> and n<sub>LacI</sub> are called "Hill coefficient". | ||
| + | α<sub>PL</sub> and α<sub>Plac</sub> depend on strength of promoter-RBS, and are adjustable. | ||
| + | Identifying value of n<sub>CI</sub> and n<sub>LacI</sub> are required for the modeling. But we know '''n<sub>CI</sub> = 3.0''' (T. Tian et al., 2006). So, we measured fluorescence intensity various IPTG concentration to identify n<sub>LacI</sub>. | ||
| - | |||
| - | + | ''' Identification of n<sub>LacI</sub> ''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | By testing how LacI represses the lac promoter, Hill coefficient of lac promoter should be decided. In order to adjust effective concentration of LacI, IPTG was added. | |
| - | In order to | + | |
| - | + | GFP fluorescence intensity was enhanced in an IPTG-dose dependent manner. It indicates that the LacI repression was getting weaker by adding IPTG. The characteristics of the lac promoter were calculated by fitting Hill function to the plots shown in figure 5-5, Finally, we obtained '''n<sub>LacI</sub> = 2.2'''. | |
| - | + | ||
| - | + | ''' Conditions for bistability''' | |
| - | + | ||
| - | + | ||
| - | + | We calculated the range of α<sub>PL</sub> in which a toggle switch model is bistability. Here, we set α<sub>Plac</sub> = 3.0. The result is below. | |
| - | |||
| - | |||
| - | |||
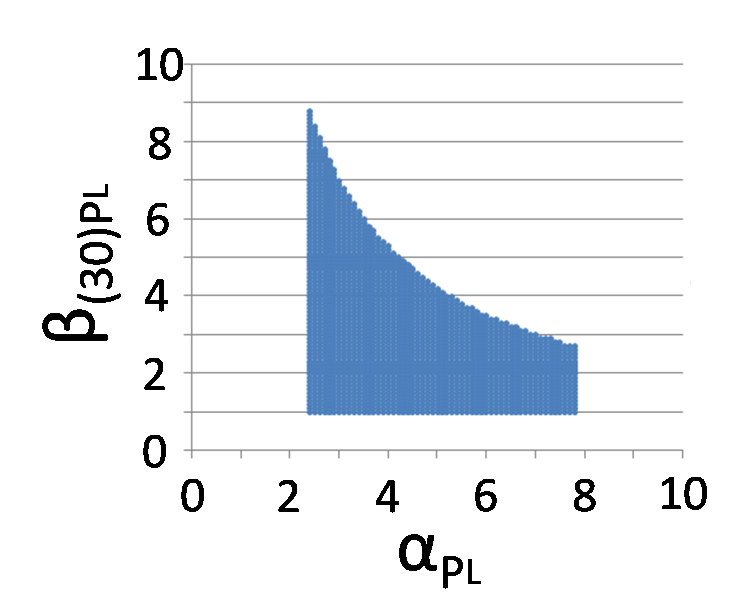
| - | + | [[Image:Conditions_for_bistability.png|300px|figure 5-5.'''P<sub>L</sub> - RBS strength range from 2.4 to 7.8''']] | |
| + | {{clear}} | ||
| - | + | ==== Pressure model ==== | |
| + | [[Image:Pressure-inducible_toggle_switch_model.png|thumb|300px|right| figure 5-6. Pressure-inducible genetic toggle switch model ]] | ||
| + | We proposed pressure-inducible genetic toggle switch model which has additional parameters to the classic toggle switch model. These parameters show the increase of activity by pressure (β<sub>(p)Plac</sub> or β<sub>(p)PL</sub>). Under atmospheric pressure (0.1 MPa), β<sub>(0.1)Plac</sub> = 1.0 and β<sub>(0.1)PL</sub> = 1.0. On the other hand, under 30 MPa, β<sub>(30)Plac</sub> = 94 and β<sub>(30)PL</sub> was not known. | ||
| + | |||
| + | In order to implement write-function, the transit of the system from bistability to monostable (P<sub>lac</sub> is stronger than P<sub>L</sub>) by 30 MPa pressure is required. Therefore, we calculated the range of β<sub>(30)PL</sub> which satisfies the above conditions. | ||
| - | |||
{{clear}} | {{clear}} | ||
| - | ==== | + | ==== The feasibility of implementation of Write/Erase cycle==== |
| - | [[Image:Desirable_condition_94.png|400px|thumb|right|figure 5- | + | [[Image:Desirable_condition_94.png|400px|thumb|right|figure 5-7. The domain of appropriate parameters if α<sub>Plac</sub> = 3.0]] |
| - | + | According to the result of simulation, we found that we can implement Write/Erase function even if P<sub>L</sub> is activated 2.5-fold when α<sub>PL</sub> = 7.8. | |
| - | + | ||
| - | + | ||
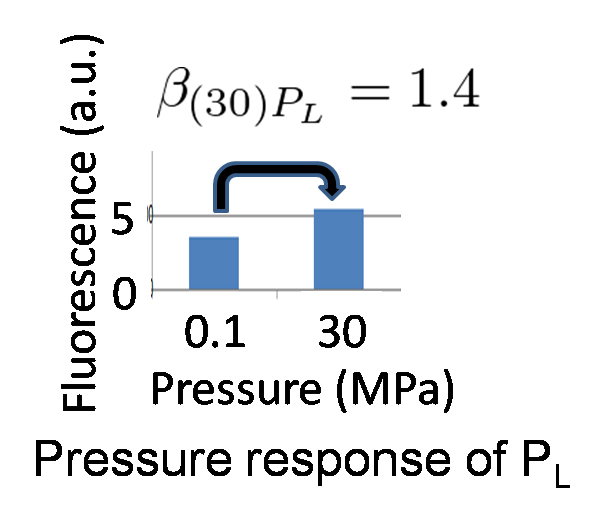
| - | [[Image:PL_pressure_response.png|200px|none|thumb|left|figure 5- | + | [[Image:PL_pressure_response.png|200px|none|thumb|left|figure 5-8. Pressure-response ability of P<sub>L</sub>]] |
| - | We identified '''β<sub> | + | We identified '''β<sub>(30)PL</sub> = 1.4''' by our wet experiment under 30MPa pressure (figure 5-6). Therefore, we can implement Write/Erase cycle if we choose an appropriate P<sub>L</sub> - RBS strength. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 05:25, 30 October 2008
| Main | Protcol | Parts Submitted to the Registry | Our Team | Acknowledgements |
|---|
Contents |
1. Our project
|
Our project is creation of "Coli Touch"!!
|

|

|
What is "Coli touch"?
“Coli Touch” has a pressure sensitive display composed of an E. coli lawn. When you touch its display, touched section is colored.
Next I'll tell you about “Coli Touch” work system. Display of “Coli Touch” contains many E. coli. When you touch this display, pressure applies to E. coli in this display. Pressure applied E. coli expresses GFP.
Why pressure?
“Coli Touch” uses pressure as input. Why do we use pressure? Past input methods (small molecules, heat and light) are difficult to induce uniformly.
Pressure can induce uniformly.
|
2. Pressure induction
Introduction
|
Tet promoters is known as a sensitive ones to pressure. (T. Sato et al., 1995) |
Construction
Result -activity of tet promoters-
|
The result shows that the tet promoter activity under 30 MPa, without repressor protein, is about 3-fold stronger than the tet promoter activity under 0.1 MPa. Therefore, we confirmed that the tet promoter was induced under 30 MPa. |
3. Touch display
Touch display (plan)
|
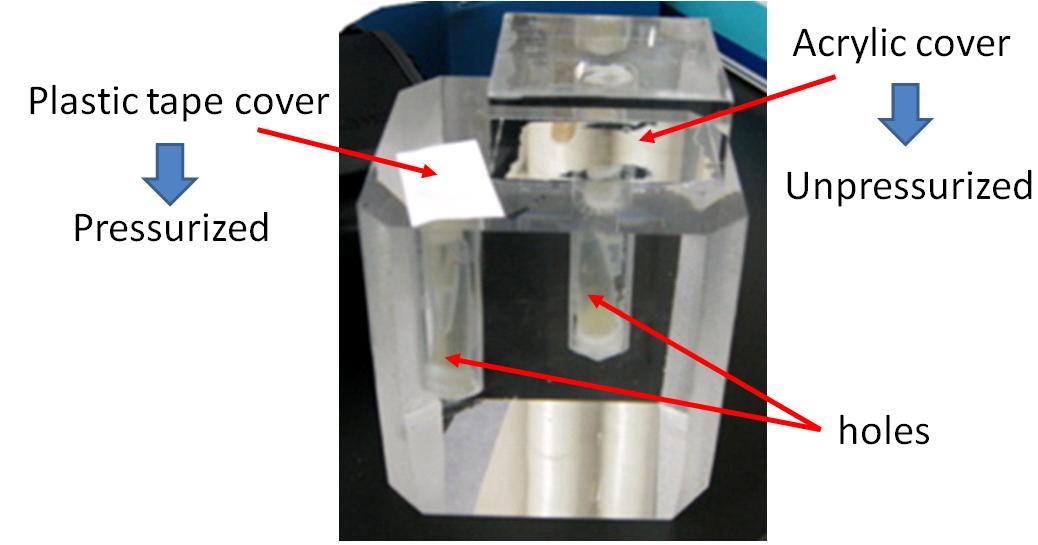
Touch display which we planned has many holes, and E. coli is in these holes.(figure 3-1 a) As the first step of creating the touch display, we created Prototype touch display. figure 3-1 b)
|
Prototype touch display
|
We created a Prototype touch display made of acrylic glasses. This touch display has two kinds of holes(show figure 3-2). Each hole contains culture medium and E. coli is cultivated in these holes. One hole (A) can be pressurized, because the hole is covered with only a plastic tape. Water pressure conducts into the hole.
The other (B) is not pressurized, because the hole is covered with a block made of acrylic glasses. Water pressure doesn’t conduct into the hole.
|
Result ~ E. coli in the touch display ~
|
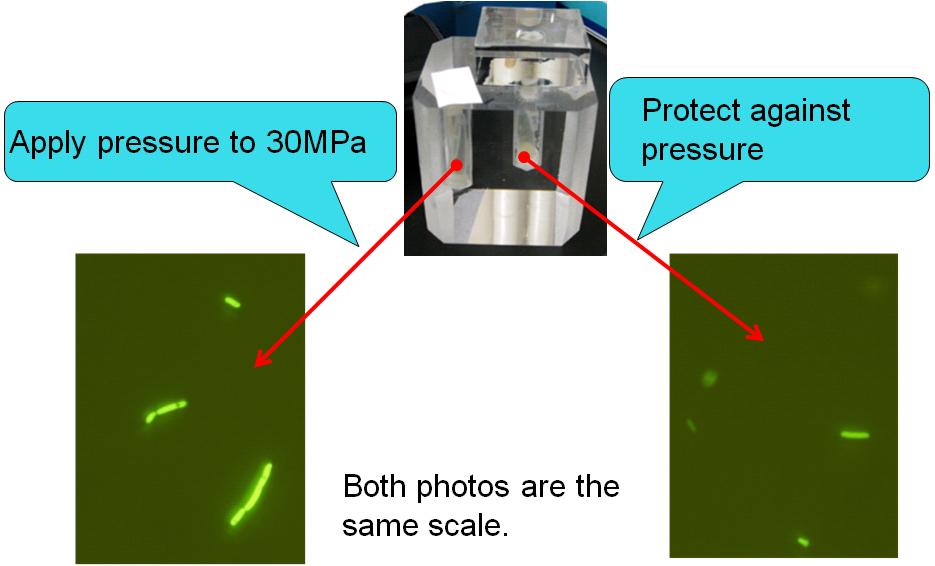
After incubation, we observed the E. coli by a fluorescence microscope. |
|
The touch display successfully regulated GFP expression in E. coli ! |
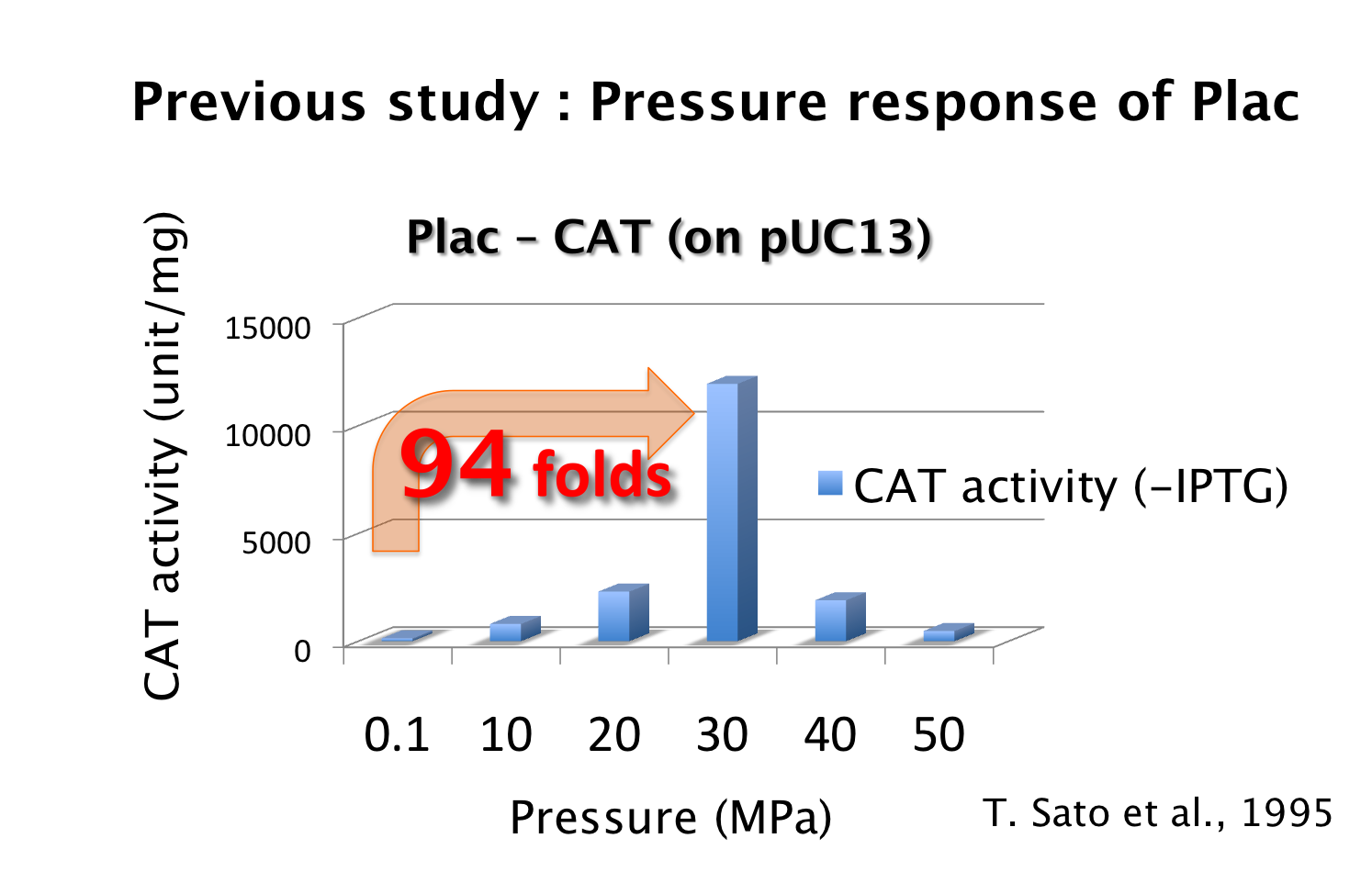
4. Low pressure-inducible promoter
Methods
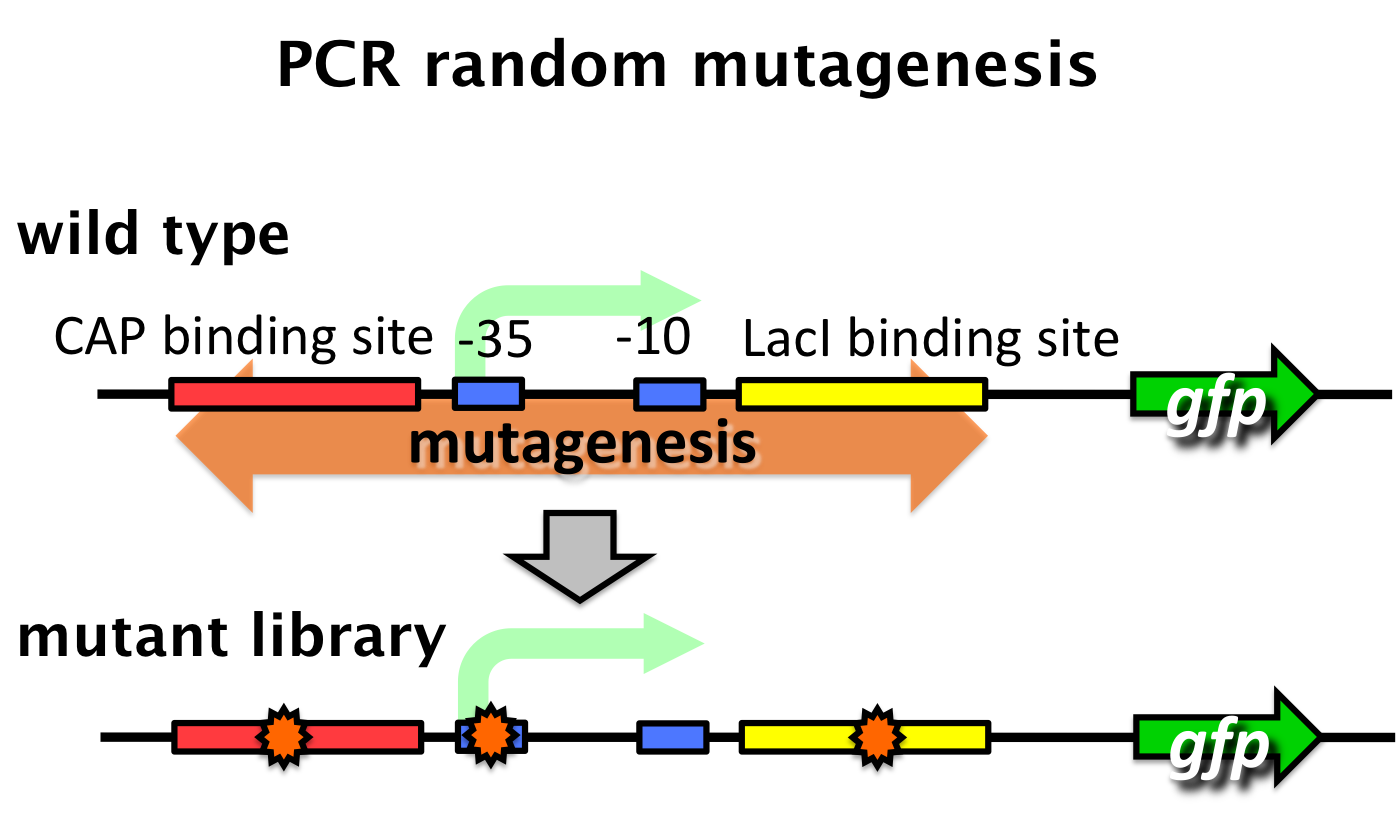
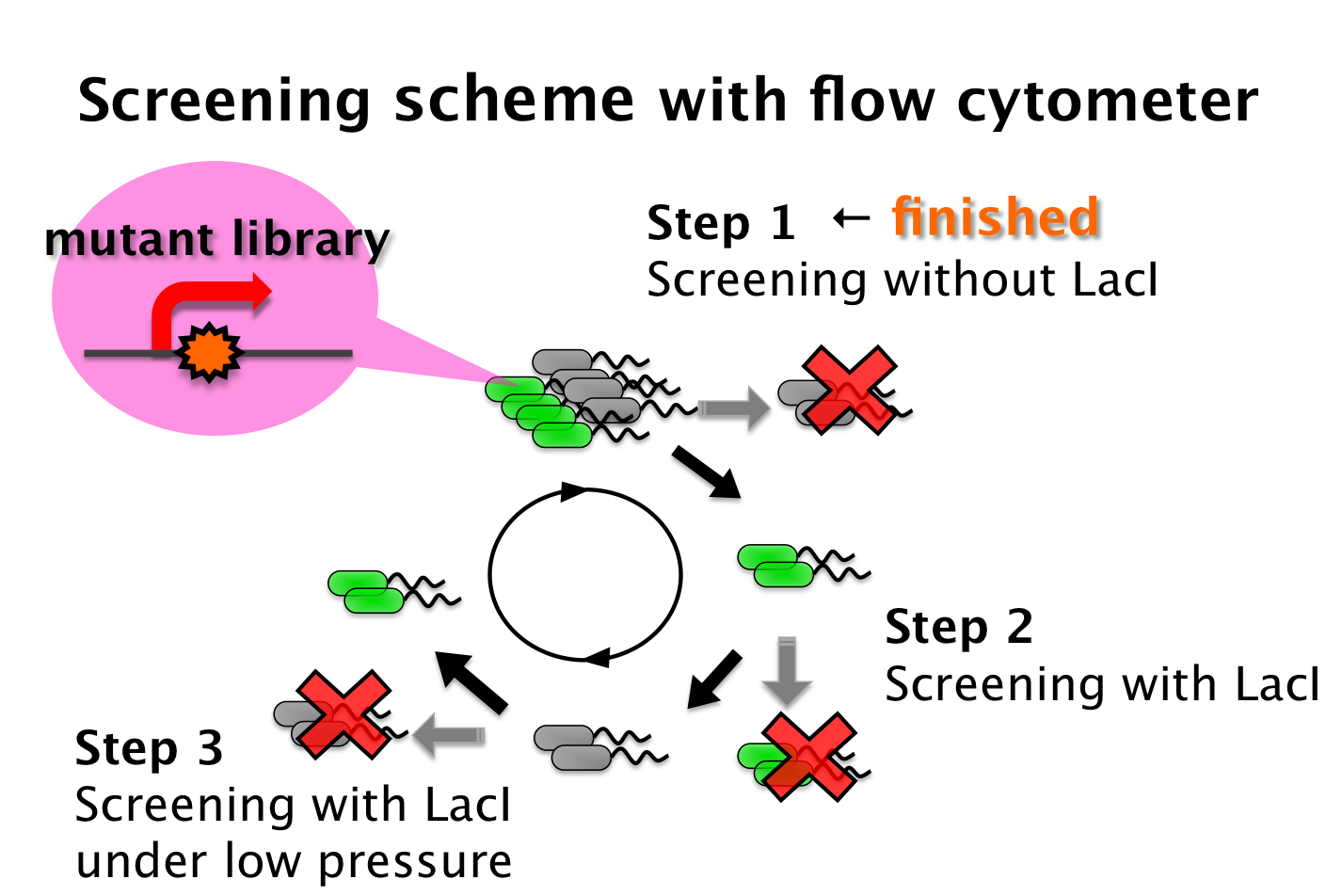
We tried to develop a low pressure inducible promoter by PCR random mutagenesis to lac promoter. Then we screened an E. coli library, with flow cytometry, for promoters that are induced under low pressure. This scheme is based on the ability to separate bacteria, with a flow cytometer, in response to expression, or lack of expression, of a fluorescent marker.
- Step 1 - Fluorescent bacteria without repressor protein were collected by a flow cytometer. This sorted pool contains bacteria bearing both constitutive and low pressure-inducible promoter.
- Step 2 - This sorted pool contains bacteria bearing both constitutive and pressure-inducible promoter fusions. False positives are removed with repressor protein. All non-fluorescent bacteria are sorted.
- Step 3 - A final passage. We sorted bacteria under pressure with repressor protein. Fluorescent bacteria are sorted. False negatives are removed. Therefore bacteria bearing promoter that are low pressure-inducible are isolated.
Results - Sequence and Characterization -
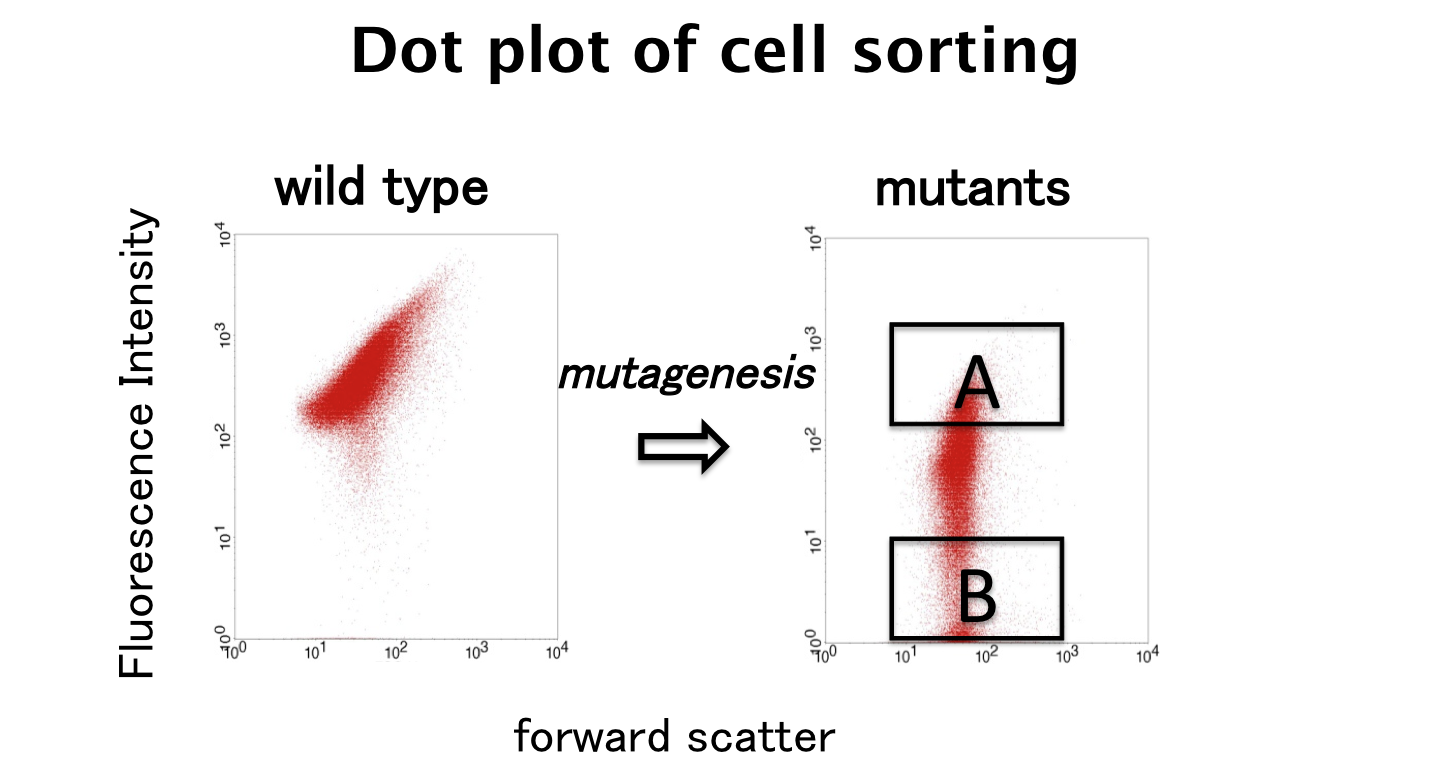
We finished step 1.
Fluorescent and non-fluorescent bacteria were sorted and we characterized their promoter.
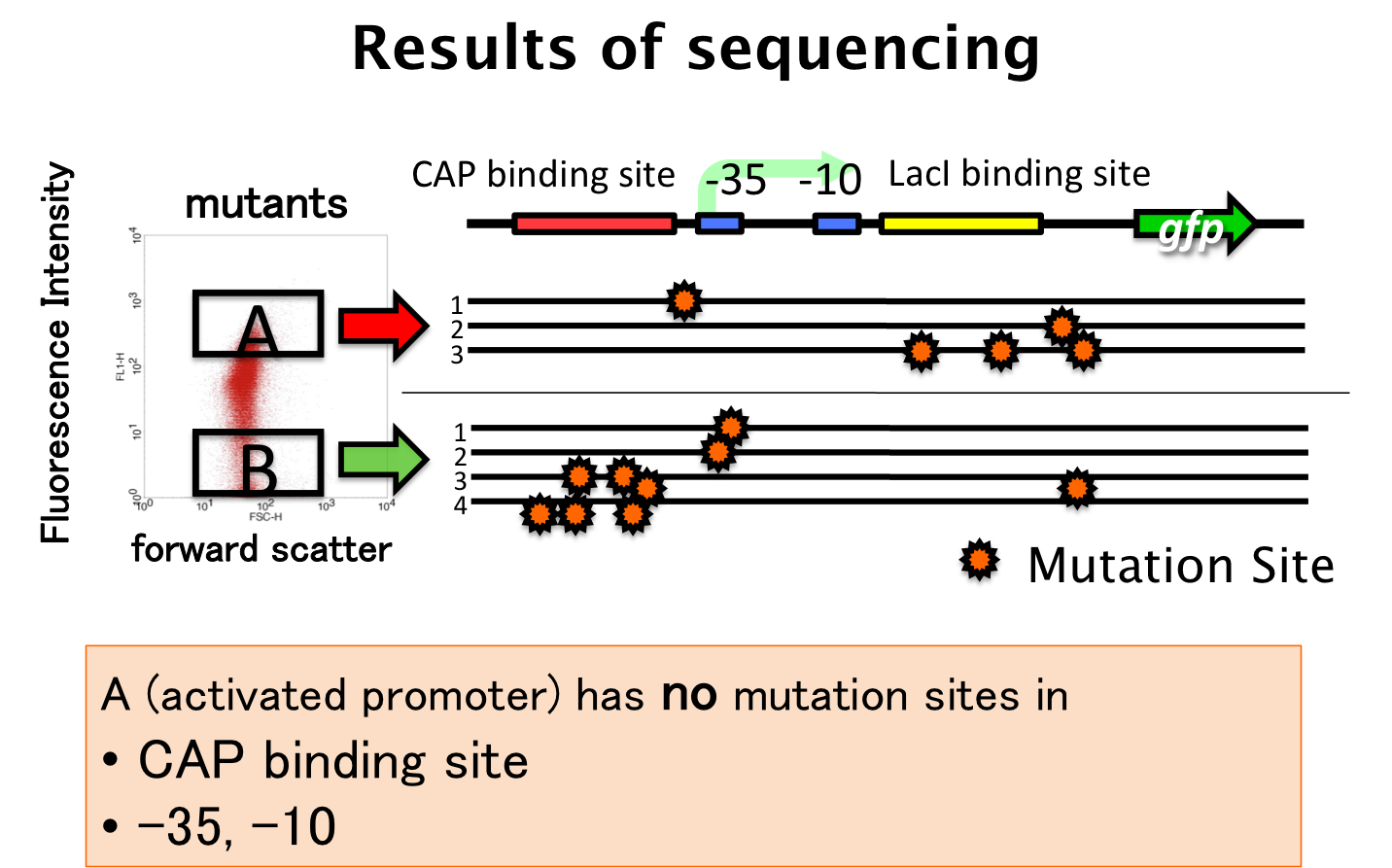
We sorted fluorescent (A) and non-fluorescent bacteria (B) with a flow cytometer. Then, we analyze these base sequences.
- A have mutations in LacI binding site or non-functional DNA.
- B have mutations in CAP binding site, -35 or non-functional DNA.
Therefore, fluorescent bacteria have no mutation in CAP binding site, -35 or -10.
We have successfully demonstrated that it is possible to collect promoters desired functions by PCR random mutagenesis and screening with a flow cytometry. This results indicate that we can screen low pressure-inducible lac promoter mutant with this methods.
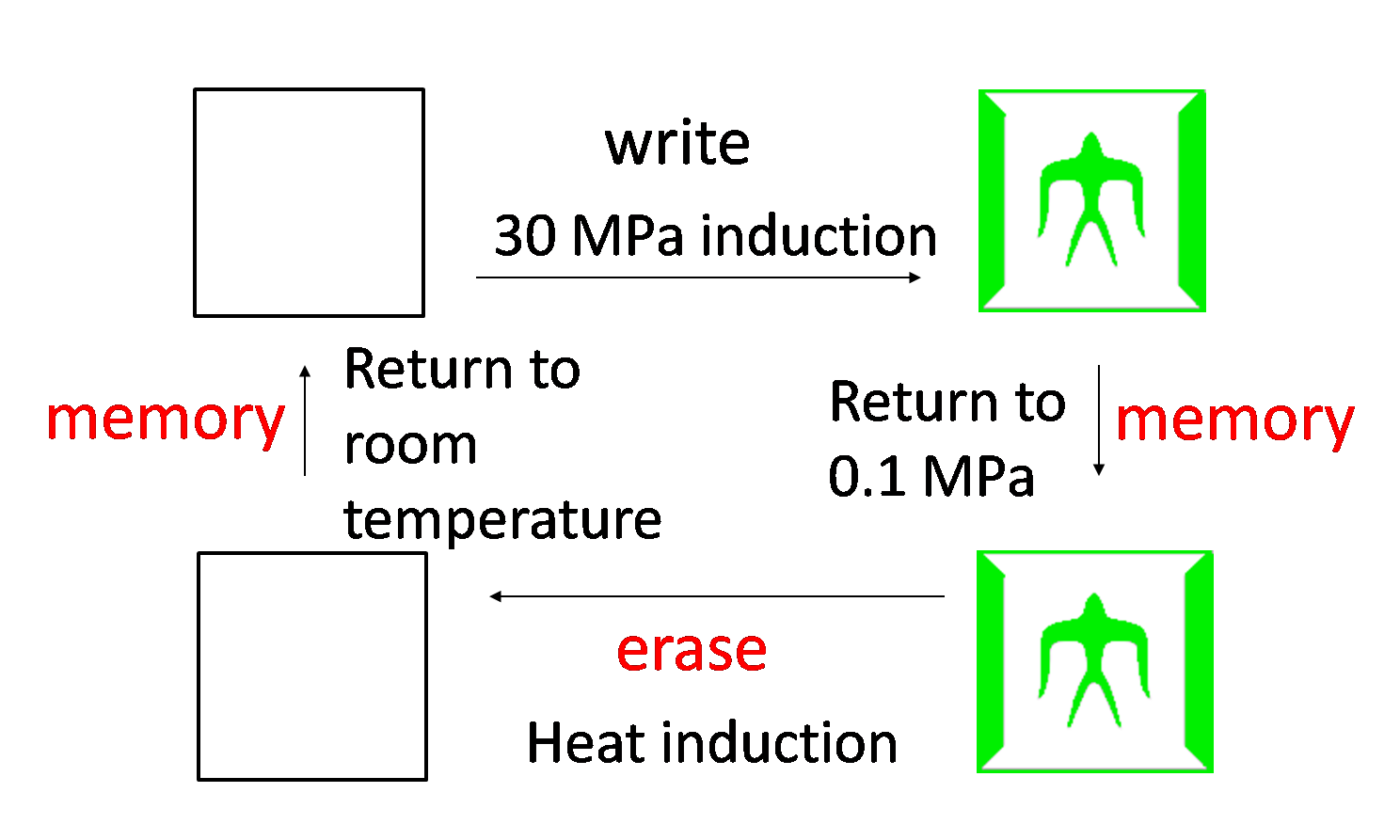
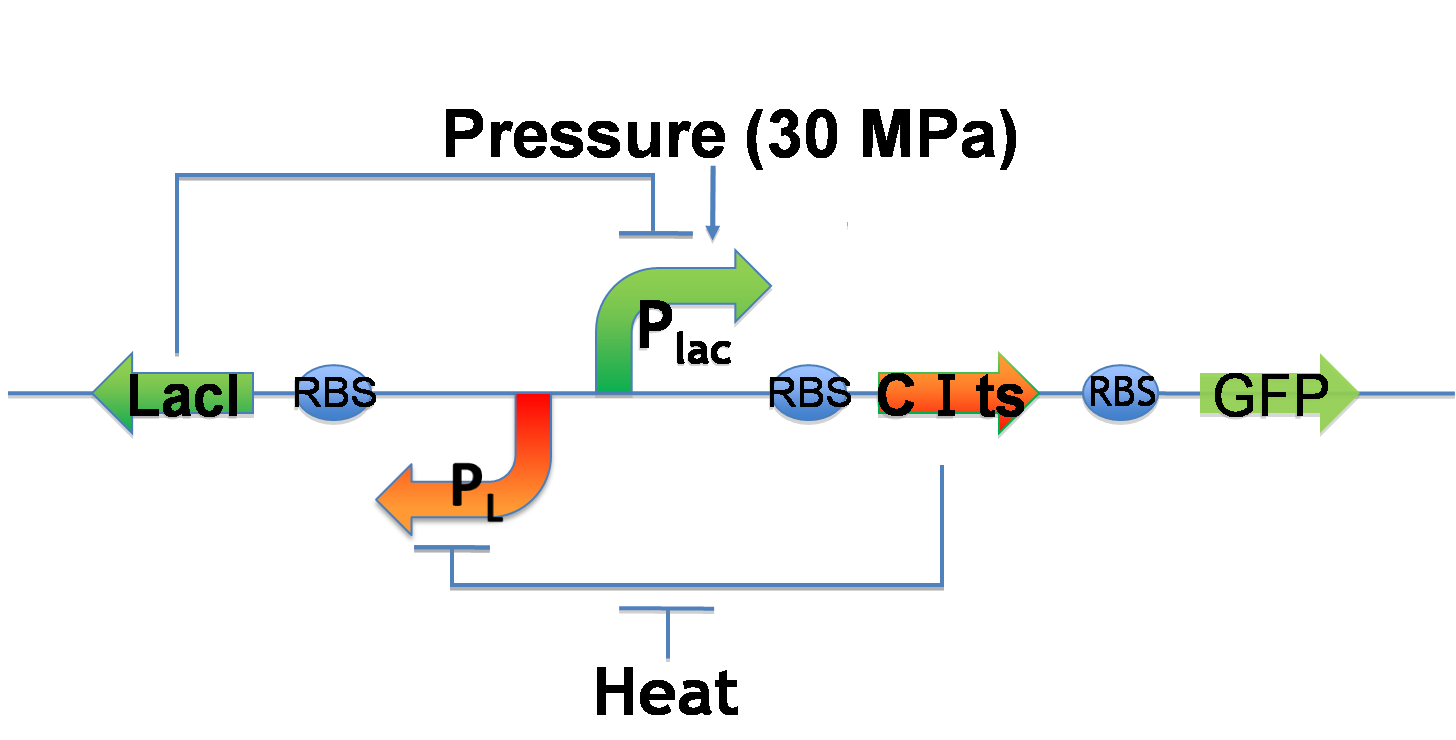
5. Write/Erase cycle
While we can implement write-function, we want to implement additionally erase-function and memory-function. Erase-function enables us to erase the painted picture, and memory-function enables us to keep the picture after we stop induction. We call these functions "Write/Erase cycle". In order to implement Write/Erase cycle, we tried to construct genetic toggle switch.
Genetic toggle switch to implement Write/Erase cycle
- Write-function
- Plac is under 30 MPa pressure.
- Plac expresses CI and GFP.
- CI represses PL and decreases LacI expression.
- Low LacI expression increases Plac activity. ⇒ Bright!!
- Erase-function
- The heat activates PL.
- PL expresses LacI.
- LacI represses Plac.
- Therefore, GFP expression decreases.
Mathematical model
Why did we use mathematical model?
As mentioned above, it is known that Plac is activated 94-fold under 30 MPa while we didn't know the increase of PL strength under 30 MPa. If PL is activated too much and Plac activity is weaker than PL activity, we can't implement write-function. So, how much is the range of the increase of PL activity under 30 MPa so as to become advantageous to that of Plac? To know this range, we need to use mathematical model.
Classical toggle switch model
Our mathematical model under 0.1 MPa is equal to a classical toggle switch model shown in figure 5-4. nCI is the cooperativity of repression of the lambda promoter, nLacI is the cooperativity of repression of the lac promoter, αPL is the effective rate of LacI synthesis and αPlac is the effective rate of CI synthesis. nCI and nLacI are called "Hill coefficient". αPL and αPlac depend on strength of promoter-RBS, and are adjustable. Identifying value of nCI and nLacI are required for the modeling. But we know nCI = 3.0 (T. Tian et al., 2006). So, we measured fluorescence intensity various IPTG concentration to identify nLacI.
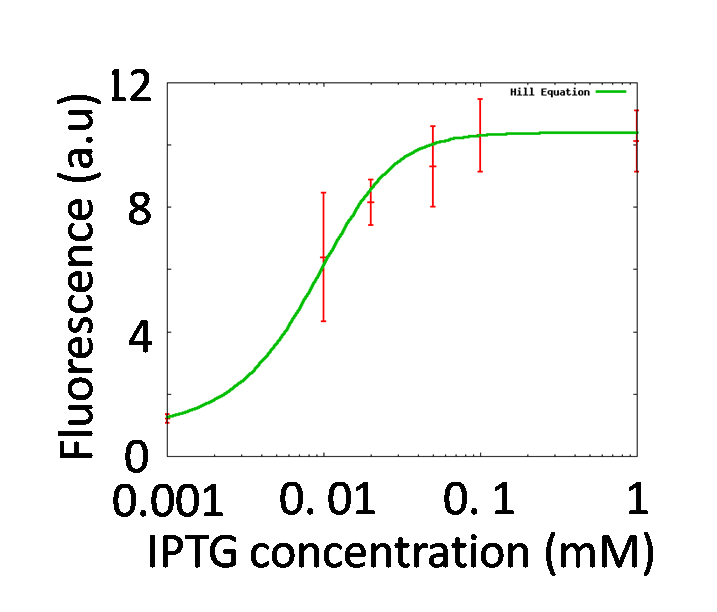
Identification of nLacI
By testing how LacI represses the lac promoter, Hill coefficient of lac promoter should be decided. In order to adjust effective concentration of LacI, IPTG was added.
GFP fluorescence intensity was enhanced in an IPTG-dose dependent manner. It indicates that the LacI repression was getting weaker by adding IPTG. The characteristics of the lac promoter were calculated by fitting Hill function to the plots shown in figure 5-5, Finally, we obtained nLacI = 2.2.
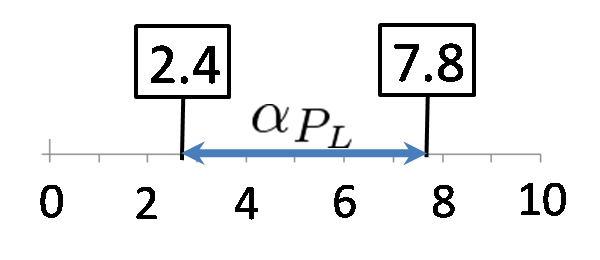
Conditions for bistability
We calculated the range of αPL in which a toggle switch model is bistability. Here, we set αPlac = 3.0. The result is below.
Pressure model
We proposed pressure-inducible genetic toggle switch model which has additional parameters to the classic toggle switch model. These parameters show the increase of activity by pressure (β(p)Plac or β(p)PL). Under atmospheric pressure (0.1 MPa), β(0.1)Plac = 1.0 and β(0.1)PL = 1.0. On the other hand, under 30 MPa, β(30)Plac = 94 and β(30)PL was not known.
In order to implement write-function, the transit of the system from bistability to monostable (Plac is stronger than PL) by 30 MPa pressure is required. Therefore, we calculated the range of β(30)PL which satisfies the above conditions.
The feasibility of implementation of Write/Erase cycle
According to the result of simulation, we found that we can implement Write/Erase function even if PL is activated 2.5-fold when αPL = 7.8.
We identified β(30)PL = 1.4 by our wet experiment under 30MPa pressure (figure 5-6). Therefore, we can implement Write/Erase cycle if we choose an appropriate PL - RBS strength.
 "
"