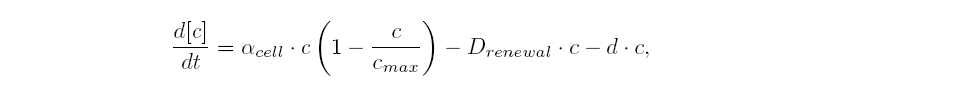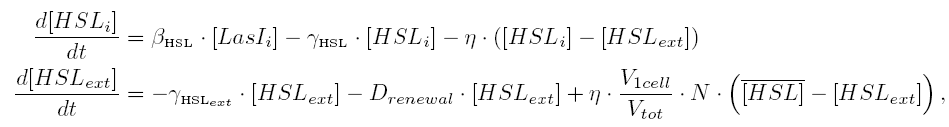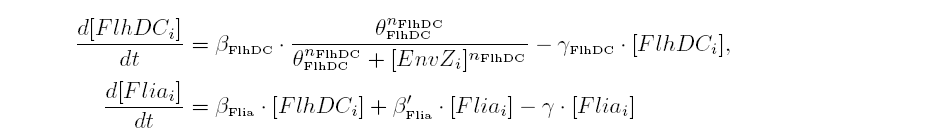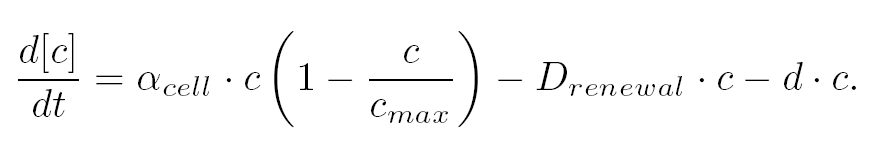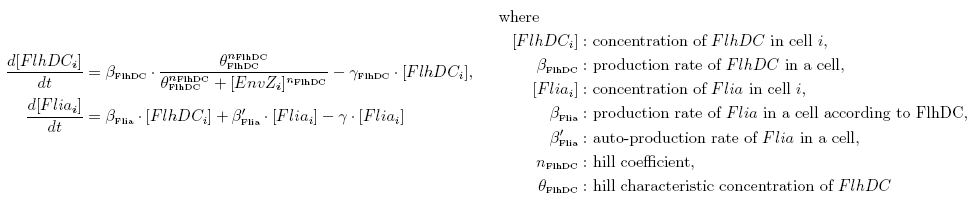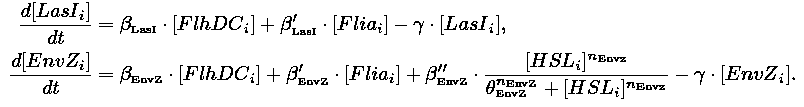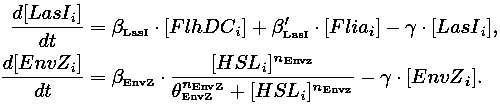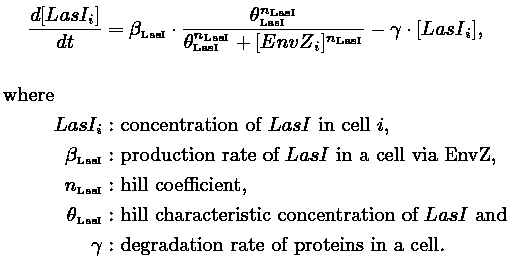Team:Paris/Analysis/Construction2
From 2008.igem.org
(→Modeling Alternatives) |
(→Introduction) |
||
| (49 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Menu}} | {{Paris/Menu}} | ||
| - | {{Paris/Header| | + | {{Paris/Header|System Improvements Description}} |
| + | {{Paris/Section_contents_analysis}} | ||
== Introduction == | == Introduction == | ||
| - | In this section, we investigate possible improvements of the [[Team:Paris/Network_analysis_and_design/Core_system/Model_construction|core system]]. Our objective is twofold : the system has to provide sustained oscillations and these oscillations should be synchronized amongst a population of cells. To this aim we explore designs inspired by quorum sensing and model in a chemostat cell growth and species diffusion outside cells. We consider two models | + | In this section, we investigate possible improvements of the [[Team:Paris/Network_analysis_and_design/Core_system/Model_construction|core system]]. Our objective is twofold : the system has to provide sustained oscillations and these oscillations should be synchronized amongst a population of cells. To this aim we explore designs inspired by quorum sensing and model in a chemostat cell growth and species diffusion outside cells. We consider two models relying on different designs principles. |
| - | {{Paris/Toggle|To | + | {{Paris/Toggle|To join our quest for alternatives click here!|Team:Paris/DescriptionMoreModelConstruction2}} |
== Modeling Alternatives == | == Modeling Alternatives == | ||
| - | The proposed | + | The proposed systems are: |
| - | + | {|align="center" cellpadding="0" cellspacing="20" | |
| - | {|align="center" | + | |
|-style="background: #D4E2EF; text-align: center;" | |-style="background: #D4E2EF; text-align: center;" | ||
| - | |''' | + | |'''HSL mediated coupled oscillators''' |
| - | |''' | + | |'''HSL mediated simple oscillator''' |
|-style="background: #ffffff; text-align: center;" | |-style="background: #ffffff; text-align: center;" | ||
| - | |[[Image:Bimo.png| | + | |[[Image:Bimo.png|400px]] |
| - | |[[Image:Unimo.png| | + | |[[Image:Unimo.png|400px]] |
|} | |} | ||
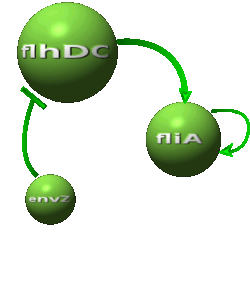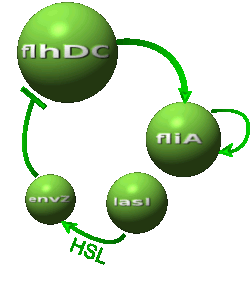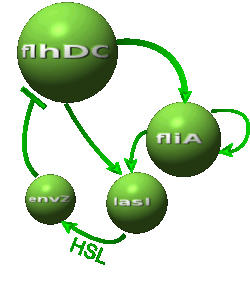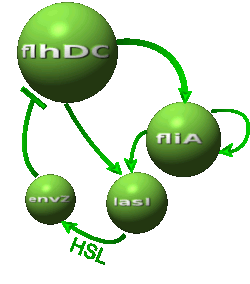
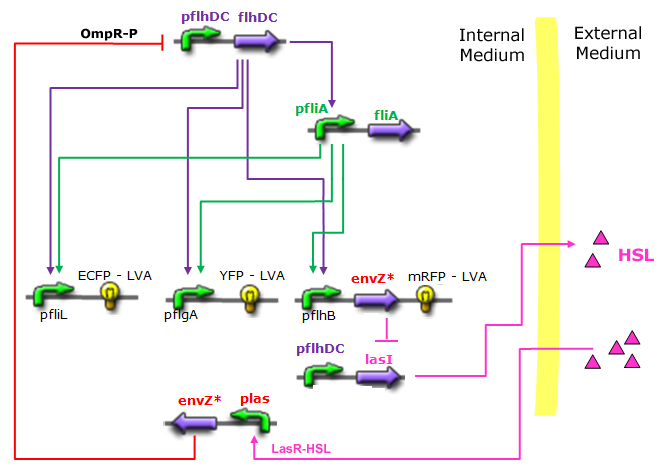
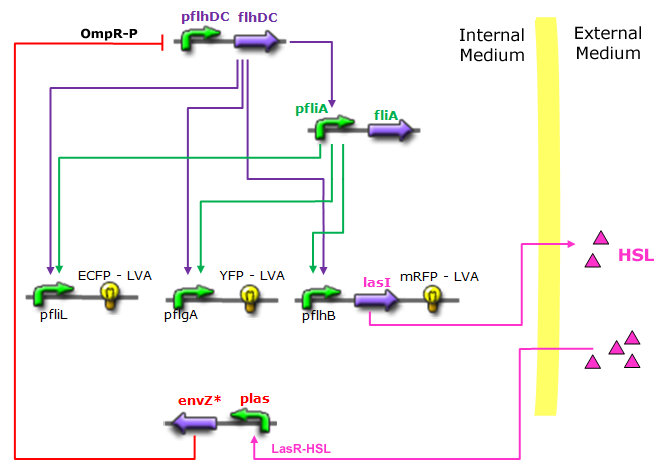
| - | *In the first system, we use a modular design. | + | *In the first system, we use a modular design. Since the core system is a 'poor' oscillator we hope that by coupling it with a 'good' oscillator we could obtain sustained oscillations in the whole system. Thus, we consider that the core system is one of the modules of the system and that the other module is a two gene oscillator system presented in [[Team:Paris/Bibliography|[2]]] that accounts for quorum sensing. We call this alternative the 'HSL mediated coupled oscillators'. |
| - | *In the second system, namely the ' | + | *In the second system, namely the 'HSL mediated simple oscillator', we rewire the architecture of the core system to introduce delay via HSL export in the environment in a single circuit. |
| - | Both the | + | Both the coupled and simple oscillators describe events that happen not only at the cellular level (as in the core system) but also at the population level due to interactions needed between a cell and its environment. |
| - | In the following sections, we first describe [[Team:Paris/Network_analysis_and_design/System_improvements/Description| | + | In the following sections, we first describe the [[Team:Paris/Network_analysis_and_design/System_improvements/Description#Common_Description|common]] part among the two proposed models and then focus our attention to the [[Team:Paris/Network_analysis_and_design/System_improvements/Description#Alternatives_Description| alternatives description]] where the characteristics that are specific to each of the modeling alternatives are presented. |
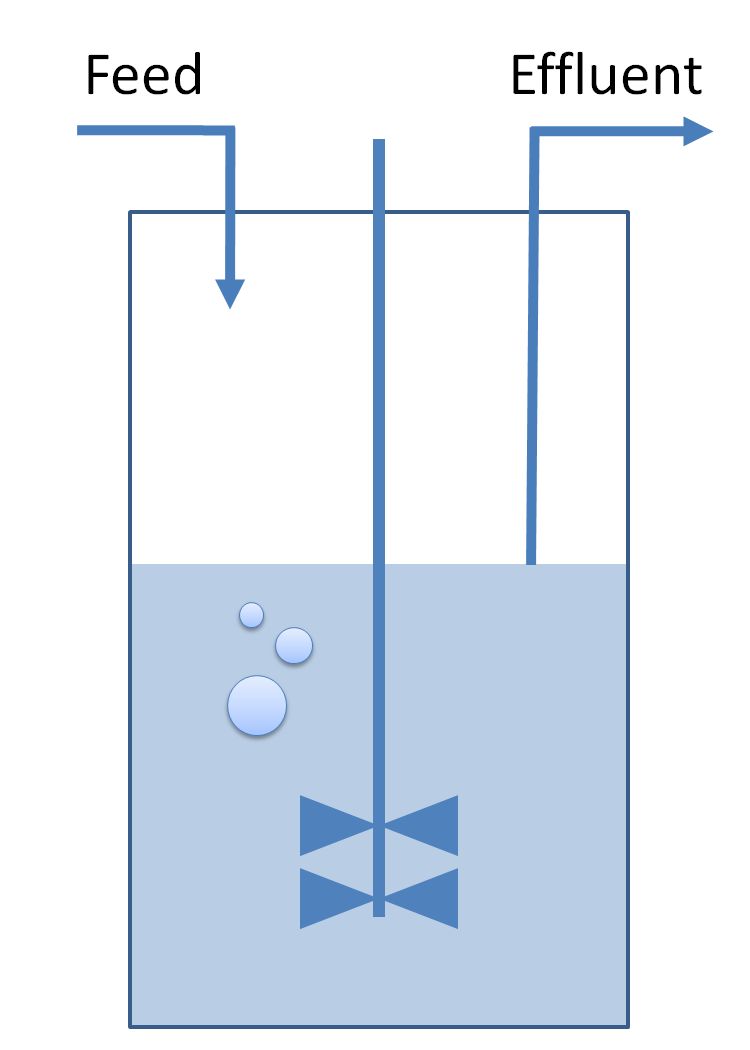
<!-- MORE DETAILS ABOUT THE CHEMOST | <!-- MORE DETAILS ABOUT THE CHEMOST | ||
* A chemostat is generally used to keep bacteria volume constant in the medium. The constant conditions provided by the chemostat help us to control bacteria growth rate. | * A chemostat is generally used to keep bacteria volume constant in the medium. The constant conditions provided by the chemostat help us to control bacteria growth rate. | ||
| Line 40: | Line 40: | ||
{|align="center" | {|align="center" | ||
|-style="background: #ffffff; text-align: left;" | |-style="background: #ffffff; text-align: left;" | ||
| - | | | + | |Logistic growth in chemostat: |
|-style="background: #ffffff; text-align: center;" | |-style="background: #ffffff; text-align: center;" | ||
|[[Image:DescriptionCommonDynamicsPart1.png|center|600px]] | |[[Image:DescriptionCommonDynamicsPart1.png|center|600px]] | ||
|-style="background: #ffffff; text-align: left;" | |-style="background: #ffffff; text-align: left;" | ||
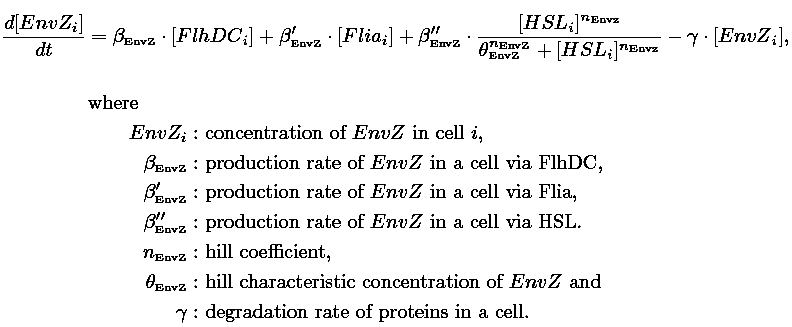
| - | | | + | |Quorum sensing by HSL diffusion: |
|-style="background: #ffffff; text-align: center;" | |-style="background: #ffffff; text-align: center;" | ||
|[[Image:DescriptionCommonDynamicsPart2.png|center|600px]] | |[[Image:DescriptionCommonDynamicsPart2.png|center|600px]] | ||
|-style="background: #ffffff; text-align: left;" | |-style="background: #ffffff; text-align: left;" | ||
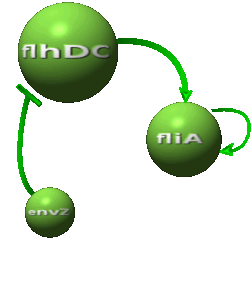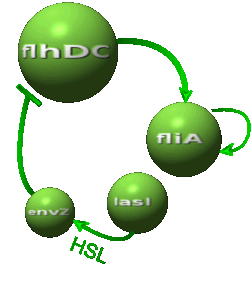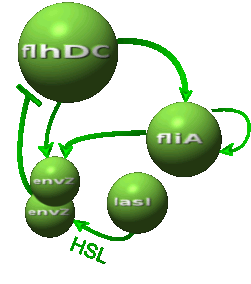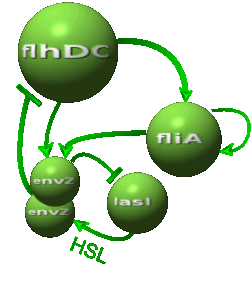
| - | | | + | |As in [[Team:Paris/Network_analysis_and_design/Core_system|core system]] flhDC inhibited by EnvZ and fliA activated by FlhDC and FliA: |
|-style="background: #ffffff; text-align: center;" | |-style="background: #ffffff; text-align: center;" | ||
|[[Image:DescriptionCommonDynamicsPart3.png|center|600px]] | |[[Image:DescriptionCommonDynamicsPart3.png|center|600px]] | ||
| Line 55: | Line 55: | ||
== Alternatives Description == | == Alternatives Description == | ||
| - | {|align="center" | + | {|align="center" cellpadding="2" cellspacing="10" |
|-style="background: #D4E2EF; text-align: center;" | |-style="background: #D4E2EF; text-align: center;" | ||
| - | |''' | + | |'''HSL mediated coupled oscillators''' |
| - | |''' | + | |'''HSL mediated simple oscillator''' |
|-style="background: #ffffff; text-align: left;" | |-style="background: #ffffff; text-align: left;" | ||
|Core system coupled with an oscillator: | |Core system coupled with an oscillator: | ||
| Line 69: | Line 69: | ||
== Kinetic parameter values== | == Kinetic parameter values== | ||
| - | + | ||
| + | Remarkably, almost all parameter values are available from experimental measurements in [[Team:Paris/Bibliography|[1]]] and from the work of Garcia-Ojalvo in [[Team:Paris/Bibliography|[2]]]. Additionally minimizing the number of parameters is possible by rescaling as done for the core system. Relevant values for the sole two missing parameters are found by exploiting a modularity assumption as described in the [[Team:Paris/Network_analysis_and_design/System_improvements/Analysis|next section]]. The following table summarize our findings: | ||
| + | |||
<center> | <center> | ||
| Line 92: | Line 94: | ||
| style="background: #dddddd;"|1 | | style="background: #dddddd;"|1 | ||
|style="background: #dddddd;"|min<sup>-1</sup> | |style="background: #dddddd;"|min<sup>-1</sup> | ||
| - | |style="background: #dddddd;"| [[Team:Paris/ | + | |style="background: #dddddd;"| [[Team:Paris/Remarks|wet-lab]] |
|- style="background: #dddddd;"| | |- style="background: #dddddd;"| | ||
| style="background: #D4E2EF;"| c<sub>max</sub> | | style="background: #D4E2EF;"| c<sub>max</sub> | ||
| Line 99: | Line 101: | ||
| style="background: #dddddd;"| 0.1 | | style="background: #dddddd;"| 0.1 | ||
| style="background: #dddddd;"| µm<sup>3</sup> | | style="background: #dddddd;"| µm<sup>3</sup> | ||
| - | | style="background: #dddddd;"| [[Team:Paris/ | + | | style="background: #dddddd;"| [[Team:Paris/Bibliography|[3]]] |
|- style="background: #dddddd;"| | |- style="background: #dddddd;"| | ||
| style="background: #D4E2EF;"| D<sub>renewal</sub> | | style="background: #D4E2EF;"| D<sub>renewal</sub> | ||
| Line 106: | Line 108: | ||
| style="background: #dddddd;"| 0.1 | | style="background: #dddddd;"| 0.1 | ||
| style="background: #dddddd;"| min<sup>-1</sup> | | style="background: #dddddd;"| min<sup>-1</sup> | ||
| - | | style="background: #dddddd;"| [[Team:Paris/ | + | | style="background: #dddddd;"| [[Team:Paris/Remarks|wet-lab]] ([[Team:Paris/Bibliography|[3]]]) |
|- style="background: #dddddd;"| | |- style="background: #dddddd;"| | ||
| style="background: #D4E2EF;"| d | | style="background: #D4E2EF;"| d | ||
| Line 113: | Line 115: | ||
| style="background: #dddddd;"| 0.5 | | style="background: #dddddd;"| 0.5 | ||
| style="background: #dddddd;"| min<sup>-1</sup> | | style="background: #dddddd;"| min<sup>-1</sup> | ||
| - | | style="background: #dddddd;"|[[Team:Paris/ | + | | style="background: #dddddd;"|[[Team:Paris/Remarks|wet-lab]] |
|- style="background: #ffffff;" | |- style="background: #ffffff;" | ||
! colspan="8" | | ! colspan="8" | | ||
| Line 131: | Line 133: | ||
| style="background: #dddddd;"|0.2690 | | style="background: #dddddd;"|0.2690 | ||
| style="background: #dddddd;"|min<sup>-1</sup> | | style="background: #dddddd;"|min<sup>-1</sup> | ||
| - | | style="background: #dddddd;"|[[Team:Paris/ | + | | style="background: #dddddd;"|[[Team:Paris/Remarks|see remarks]] |
|- style="background: #dddddd;"| | |- style="background: #dddddd;"| | ||
| style="background: #D4E2EF;"|γ<sub>HSL<sub>ext</sub></sub> <br> <br> | | style="background: #D4E2EF;"|γ<sub>HSL<sub>ext</sub></sub> <br> <br> | ||
| Line 138: | Line 140: | ||
| style="background: #dddddd;"|0.5380 | | style="background: #dddddd;"|0.5380 | ||
|style="background: #dddddd;"| min<sup>-1</sup> | |style="background: #dddddd;"| min<sup>-1</sup> | ||
| - | | style="background: #dddddd;"|[[Team:Paris/ | + | | style="background: #dddddd;"|[[Team:Paris/Bibliography|[2]]] |
|- style="background: #dddddd;"| | |- style="background: #dddddd;"| | ||
| style="background: #D4E2EF;" |β<sub>HSL</sub> | | style="background: #D4E2EF;" |β<sub>HSL</sub> | ||
| Line 145: | Line 147: | ||
| style="background: #dddddd;"|16 | | style="background: #dddddd;"|16 | ||
| style="background: #dddddd;"|min<sup>-1</sup> | | style="background: #dddddd;"|min<sup>-1</sup> | ||
| - | | style="background: #dddddd;"|[[Team:Paris/ | + | | style="background: #dddddd;"|[[Team:Paris/Bibliography|∅]] |
|- style="background: #dddddd;"| | |- style="background: #dddddd;"| | ||
| style="background: #D4E2EF;" |η | | style="background: #D4E2EF;" |η | ||
| Line 152: | Line 154: | ||
| style="background: #dddddd;"|505 | | style="background: #dddddd;"|505 | ||
| style="background: #dddddd;"|min<sup>-1</sup> | | style="background: #dddddd;"|min<sup>-1</sup> | ||
| - | | style="background: #dddddd;"|[[Team:Paris/ | + | | style="background: #dddddd;"|[[Team:Paris/Bibliography|[2]]] |
|- style="background: #dddddd;"| | |- style="background: #dddddd;"| | ||
| style="background: #D4E2EF;" |n<sub>HSL</sub> | | style="background: #D4E2EF;" |n<sub>HSL</sub> | ||
| Line 159: | Line 161: | ||
| style="background: #dddddd;"|4 | | style="background: #dddddd;"|4 | ||
| style="background: #dddddd;"| | | style="background: #dddddd;"| | ||
| - | | style="background: #dddddd;"|[[Team:Paris/ | + | | style="background: #dddddd;"|[[Team:Paris/Bibliography|[3]]] |
|- style="background: #dddddd;"| | |- style="background: #dddddd;"| | ||
| style="background: #D4E2EF;" |θ<sub>HSL</sub> | | style="background: #D4E2EF;" |θ<sub>HSL</sub> | ||
| Line 165: | Line 167: | ||
| style="background: #dddddd;"| | | style="background: #dddddd;"| | ||
| style="background: #dddddd;"|0.5 | | style="background: #dddddd;"|0.5 | ||
| - | | style="background: #dddddd;"|[[Team:Paris/ | + | | style="background: #dddddd;"|[[Team:Paris/Remarks|c.u.]] |
| - | | style="background: #dddddd;"|[[Team:Paris/ | + | | style="background: #dddddd;"|[[Team:Paris/Bibliography|[3]]] |
|}</center> | |}</center> | ||
| + | |||
| + | |||
| + | {{Paris/Navig|Team:Paris/Analysis}} | ||
Latest revision as of 03:18, 30 October 2008
|
System Improvements Description
Other pages:
IntroductionIn this section, we investigate possible improvements of the core system. Our objective is twofold : the system has to provide sustained oscillations and these oscillations should be synchronized amongst a population of cells. To this aim we explore designs inspired by quorum sensing and model in a chemostat cell growth and species diffusion outside cells. We consider two models relying on different designs principles. ↓ To join our quest for alternatives click here! ↑
Modeling AlternativesThe proposed systems are:
Both the coupled and simple oscillators describe events that happen not only at the cellular level (as in the core system) but also at the population level due to interactions needed between a cell and its environment. In the following sections, we first describe the common part among the two proposed models and then focus our attention to the alternatives description where the characteristics that are specific to each of the modeling alternatives are presented. Common Description
↓ read more on common dynamics... ↑
Alternatives Description
↓ read more on alternatives description... ↑
Kinetic parameter valuesRemarkably, almost all parameter values are available from experimental measurements in [1] and from the work of Garcia-Ojalvo in [2]. Additionally minimizing the number of parameters is possible by rescaling as done for the core system. Relevant values for the sole two missing parameters are found by exploiting a modularity assumption as described in the next section. The following table summarize our findings:
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"