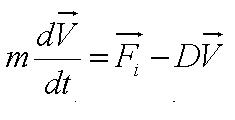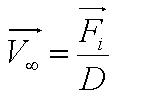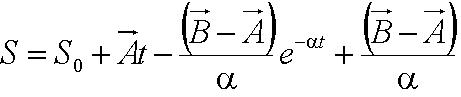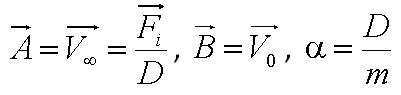Team:Imperial College/Motility
From 2008.igem.org
m |
|||
| (7 intermediate revisions not shown) | |||
| Line 23: | Line 23: | ||
===== Data Extraction ===== | ===== Data Extraction ===== | ||
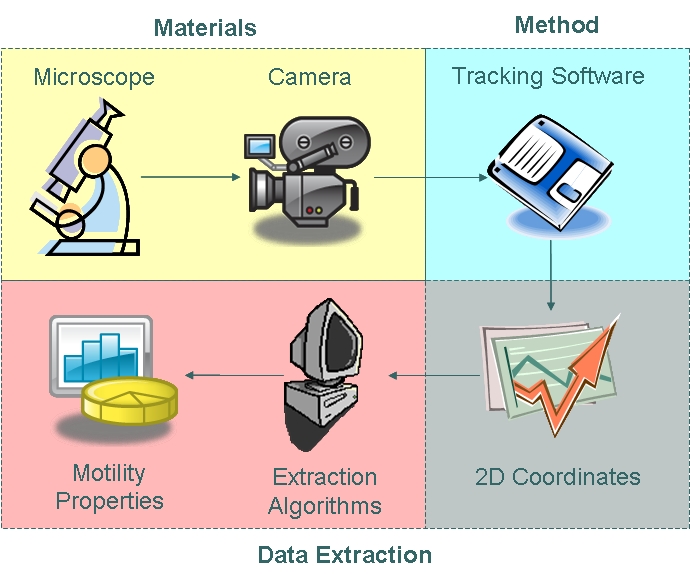
| - | The coordinate data obtained were then fed into algorithms to model cell trajectory and motility. Algorithms used to extract motility data and fit cell trajectory data to models can be found in the | + | The coordinate data obtained were then fed into algorithms to model cell trajectory and motility. Algorithms used to extract motility data and fit cell trajectory data to models can be found in the appendices section.|}} |
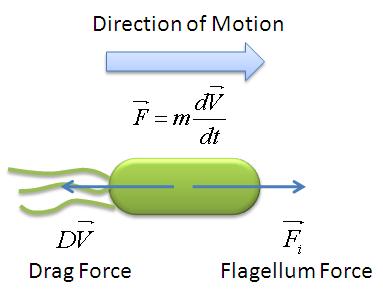
{{Imperial/Box1|Motility Model| | {{Imperial/Box1|Motility Model| | ||
| Line 54: | Line 54: | ||
<br> | <br> | ||
Given the random motion of bacteria, the outcome to such modelling will be the probability distribution of the model parameters and the time between changes of flagellar force. | Given the random motion of bacteria, the outcome to such modelling will be the probability distribution of the model parameters and the time between changes of flagellar force. | ||
| - | |||
|}} | |}} | ||
| - | {{Imperial/ | + | {{Imperial/Box1|Results| |
| - | + | ||
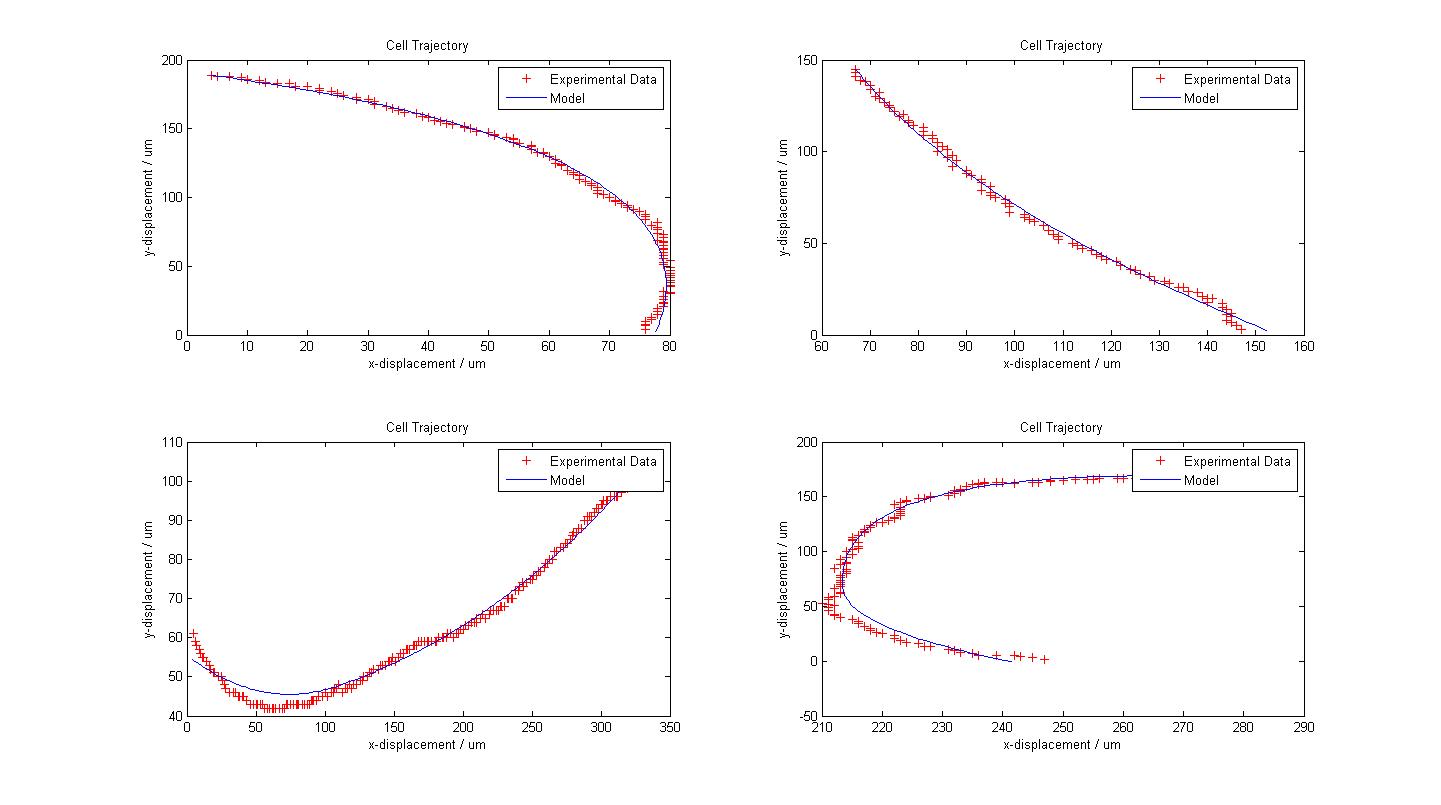
In the course of our project, we mainly focused on obtaining the flagellar force, which is associated with the parameter A. <br> | In the course of our project, we mainly focused on obtaining the flagellar force, which is associated with the parameter A. <br> | ||
| Line 76: | Line 74: | ||
{{Pipe}}- | {{Pipe}}- | ||
{{Pipe}}} | {{Pipe}}} | ||
| - | |||
| - | |||
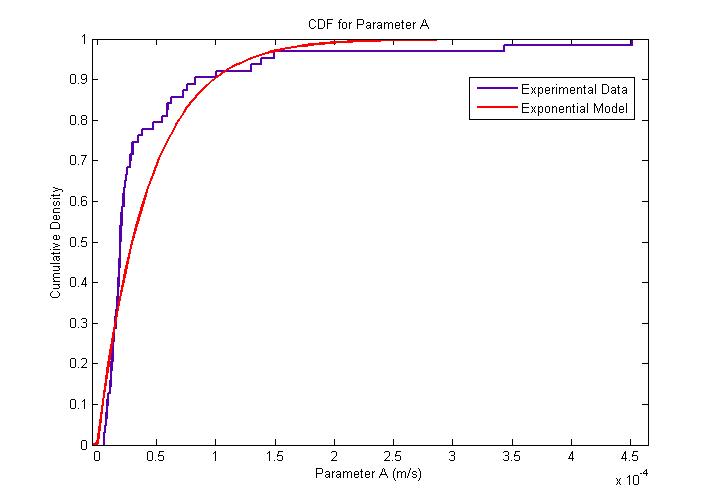
The MATLAB Distribution Fitting Tool was used to model the distribution of the amplitude of parameter ''A''. | The MATLAB Distribution Fitting Tool was used to model the distribution of the amplitude of parameter ''A''. | ||
<br> | <br> | ||
| Line 96: | Line 92: | ||
|}} | |}} | ||
| - | {{Imperial/ | + | {{Imperial/Box2|Conclusion| |
| - | From our model fitting process, we can see that flagellar force is exponentially distributed. Our mechanical model though simple, fits the cell trajectory data extremely well as shown in the figure above. Further work would involve the use of a movable stage to track the movement of ''B. subtilis'' over its entire run, so as to obtain a distribution of other motility parameters associated with running and tumbling events. | + | From our model fitting process, we can see that flagellar force is exponentially distributed. Our mechanical model though simple, fits the cell trajectory data extremely well as shown in the figure above. Further work would involve the use of a movable stage to track the movement of ''B. subtilis'' over its entire run, so as to obtain a distribution of other motility parameters associated with running and tumbling events. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ===== References ===== | ||
| + | #{{FormatRef|H C Berg |1975|Chemotaxis in Bacteria|Annual Review of Biophysics and Bioengineering Vol.4|119-136||}} | ||
| + | <br>|}} | ||
{{Imperial/EndPage|Genetic Circuit|Appendices}} | {{Imperial/EndPage|Genetic Circuit|Appendices}} | ||
Latest revision as of 03:14, 30 October 2008
Motility Analysis
|
|||||||||||||||||||||||||
 "
"