DNA-Origami
From 2008.igem.org
(Difference between revisions)
m |
|||
| (5 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<br><br> | <br><br> | ||
<h2>Introduction</h2> | <h2>Introduction</h2> | ||
| + | [[Image:Freiburg2008_Fab_on_Origami_animated.gif|right|400 px]] | ||
Paul Rothemund has discovered that it is possible to shape M13-Phage single-strand-DNA simply adding oligonucleotides that will work as „brackets“ when complementing the long single-strand.<br> In this way, one can generate for example DNA-squares of a certain size with „nods“ at certain distances. | Paul Rothemund has discovered that it is possible to shape M13-Phage single-strand-DNA simply adding oligonucleotides that will work as „brackets“ when complementing the long single-strand.<br> In this way, one can generate for example DNA-squares of a certain size with „nods“ at certain distances. | ||
One member of our team, Daniel Hautzinger, has recently finished his diploma-thesis on Origami-DNA and the possibilities of generating patterns on these square surfaces by modifying the oligo-nucleotides that build up the nod-points.<br> | One member of our team, Daniel Hautzinger, has recently finished his diploma-thesis on Origami-DNA and the possibilities of generating patterns on these square surfaces by modifying the oligo-nucleotides that build up the nod-points.<br> | ||
As the antigens NIP and fluoresceine can as well be fused to these oligos, we had found the seemingly perfect tool to present strictly defined two-dimensional antigen-patterns to cells carrying our synthetic receptor system. | As the antigens NIP and fluoresceine can as well be fused to these oligos, we had found the seemingly perfect tool to present strictly defined two-dimensional antigen-patterns to cells carrying our synthetic receptor system. | ||
| - | + | ||
<h2>Methods</h2> | <h2>Methods</h2> | ||
<h3>Phage DNA<span style="font-weight: bold;"></span></h3> | <h3>Phage DNA<span style="font-weight: bold;"></span></h3> | ||
| Line 38: | Line 39: | ||
gently by inverting the tube. The mixture (solution 3) was left for 1 h | gently by inverting the tube. The mixture (solution 3) was left for 1 h | ||
on ice. Solution 3 was centrifuged at 13200 rpm for 10 min.<br> | on ice. Solution 3 was centrifuged at 13200 rpm for 10 min.<br> | ||
| - | <h4> | + | <h4>Measurement of phage titers</h4> |
The absorption of solution 3 was measured on a Jasco V-550 UV/VIS | The absorption of solution 3 was measured on a Jasco V-550 UV/VIS | ||
spectrometer at 269 nm. <br> | spectrometer at 269 nm. <br> | ||
| Line 44: | Line 45: | ||
<span style="font-style: italic;"><span | <span style="font-style: italic;"><span | ||
style="font-weight: bold;">Phage DNA</span> = | style="font-weight: bold;">Phage DNA</span> = | ||
| - | ((A269- | + | ((A269-A320) * 6 * 10^16 * dilution factor) / (number of bases in the |
phage genom = 7249 bp)</span><br> | phage genom = 7249 bp)</span><br> | ||
<h4>Isolation of the phage DNA </h4> | <h4>Isolation of the phage DNA </h4> | ||
| Line 50: | Line 51: | ||
(Cat.No: 27704). <br> | (Cat.No: 27704). <br> | ||
DNA-concentration was quantified by Nano-drop photometer.<br> | DNA-concentration was quantified by Nano-drop photometer.<br> | ||
| - | <span style="font-weight: bold;"><br> | + | <span style="font-weight: bold;"><br></span> |
| - | </span> | + | |
<h3><span style="font-weight: bold;">Origami</span></h3> | <h3><span style="font-weight: bold;">Origami</span></h3> | ||
| - | <h4><span style="font-weight: bold;"></span> | + | |
| - | + | <h4><span style="font-weight: bold;"></span>Origami production<br></h4> | |
| - | </h4> | + | |
To produce the Origami we mixed each the M13mp18 DNA with the oligos, | To produce the Origami we mixed each the M13mp18 DNA with the oligos, | ||
water and TEA/MgAcetat (end concentration =12.5mM).<br> | water and TEA/MgAcetat (end concentration =12.5mM).<br> | ||
| Line 71: | Line 72: | ||
the 2 oligos with the Alexa 488 were used.<br> | the 2 oligos with the Alexa 488 were used.<br> | ||
<br> | <br> | ||
| - | <h4><span style="font-weight: bold;"></span>Purification of | + | <h4><span style="font-weight: bold;"></span>Purification of DNA-Origamis<br> |
</h4> | </h4> | ||
To purify the DNA-Origamis from the unbound DNA-oligos, we used Montage® PCR Centrifugal Filter Devices (Millipore). The Montage® PCR Centrifugal Filter Devices were labeled and put with the purple side on top in 1.5 ml Eppendorf tubes. To clean the filter of remaining glycerol, 450 µl TAE/MgAcetat (12.5 mM; 1x filtered) was put on top of the filter and centrifuged for 15 min at 1000 g. After removing the filtrate, 400 µl TEA/MgAcetat (12.5 mM;1x filtered) and 45 µl DNA-origami were put on top of the filter and again centrifuged for 15 min at 1000 g. The filtrate was removed again. All unbound DNA-oligos were washed off by putting 400 µl TEA/MgAcetat (12.5 mM; 1x filtered) on top of the filter. The sample was centrifuged for 15 min at 1000 g. To release the DNA-origamis of the filter, 100 µl TAE/MgAcetat (12.5 mM;1x filtered) was put on top of the filter, and the filter was left at room temperature for at least 2 min. The filter shouldn´t run dry. The Montage® PCR Centrifugal Filter Devices were put upside down (the purple side has to be on the bottom) in one of the special Invert Spin tubes form Millipore and centrifuged for 3 min at 1000 g. | To purify the DNA-Origamis from the unbound DNA-oligos, we used Montage® PCR Centrifugal Filter Devices (Millipore). The Montage® PCR Centrifugal Filter Devices were labeled and put with the purple side on top in 1.5 ml Eppendorf tubes. To clean the filter of remaining glycerol, 450 µl TAE/MgAcetat (12.5 mM; 1x filtered) was put on top of the filter and centrifuged for 15 min at 1000 g. After removing the filtrate, 400 µl TEA/MgAcetat (12.5 mM;1x filtered) and 45 µl DNA-origami were put on top of the filter and again centrifuged for 15 min at 1000 g. The filtrate was removed again. All unbound DNA-oligos were washed off by putting 400 µl TEA/MgAcetat (12.5 mM; 1x filtered) on top of the filter. The sample was centrifuged for 15 min at 1000 g. To release the DNA-origamis of the filter, 100 µl TAE/MgAcetat (12.5 mM;1x filtered) was put on top of the filter, and the filter was left at room temperature for at least 2 min. The filter shouldn´t run dry. The Montage® PCR Centrifugal Filter Devices were put upside down (the purple side has to be on the bottom) in one of the special Invert Spin tubes form Millipore and centrifuged for 3 min at 1000 g. | ||
The Origami were kept in different buffers. For this, TEA/MgAcetat (12.5 mM; 1x filtered) was replaced by the according buffer.<br> | The Origami were kept in different buffers. For this, TEA/MgAcetat (12.5 mM; 1x filtered) was replaced by the according buffer.<br> | ||
| - | <h4><span style="font-weight: bold;"></span>Atomic force microscopy to | + | <h4><span style="font-weight: bold;"></span>Atomic force microscopy to test the origami stability |
<br> | <br> | ||
</h4> | </h4> | ||
| Line 82: | Line 83: | ||
The metal pane was fixed in the metal sample holder by a magnet, so that the sample could not move itself during the measurement. | The metal pane was fixed in the metal sample holder by a magnet, so that the sample could not move itself during the measurement. | ||
<h2>Results and Discussion</h2> | <h2>Results and Discussion</h2> | ||
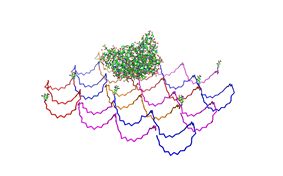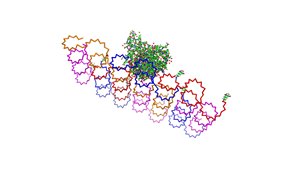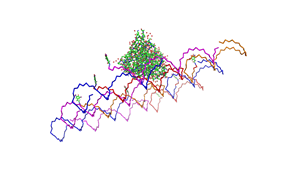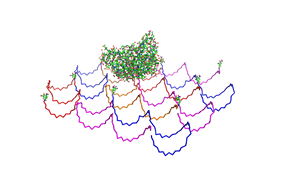
| - | <h3>1. Different ratios of | + | <h3>1. Different ratios of phage DNA to staple oligonucleotides</h3> |
Because we also wanted to measure the calcium influx in the LSRII | Because we also wanted to measure the calcium influx in the LSRII | ||
fluorescence spectrometer, we had to increase the concentration of the | fluorescence spectrometer, we had to increase the concentration of the | ||
Origamis at least up to 200 nM. Because the oligos we had to use for | Origamis at least up to 200 nM. Because the oligos we had to use for | ||
| - | building | + | building DNA-Origami are very expensive, we first tried to reduce the |
| - | ratio of | + | ratio of phage DNA to oligos. Therefore we tried to make Origamis with two |
(1:10) and four (1:5) times lower concentration of oligos. As positive | (1:10) and four (1:5) times lower concentration of oligos. As positive | ||
control we took the 1:20 ratio at which we knew it should be stable. We | control we took the 1:20 ratio at which we knew it should be stable. We | ||
| - | used | + | used AFM to check if the Origami are well formed. The results are |
shown in figures 1-3. <br> | shown in figures 1-3. <br> | ||
<br> | <br> | ||
| Line 140: | Line 141: | ||
of the Origami in phosphate buffer without calcium should be tested.<br> | of the Origami in phosphate buffer without calcium should be tested.<br> | ||
<br> | <br> | ||
| - | <h2>Literature</h2> | + | <h2>[[Image:MO2.jpg|50px|]]Literature</h2> |
| - | *Paul W. K. Rothemund: Nature 440, 297-302 (16 March 2006) | + | *Paul W. K. Rothemund:"Folding DNA to create nanoscale shapes and patterns", Nature 440, 297-302 (16 March 2006) |
}} | }} | ||
Latest revision as of 01:50, 30 October 2008
 "
"