Team:Chiba/Experiments:copy number
From 2008.igem.org
(→Design) |
(→Experiment) |
||
| Line 36: | Line 36: | ||
===Experiment=== | ===Experiment=== | ||
| - | + | We used a pMB1 ori high copy vector containing BBa_T9002 and a low | |
| + | copy vector containing BBa_T9002 with a P15A as ori. | ||
| - | + | We determined the effects of altering the plasmid copy number by | |
| + | comparing the above situations. | ||
| - | + | Experiments used the following genetic circuits: | |
| + | <br clear=all> | ||
{| class="tbl" | | {| class="tbl" | | ||
|- | |- | ||
| Line 61: | Line 64: | ||
<br clear=all> | <br clear=all> | ||
| - | + | In addition, a Chloramphenicol resistance marker-containing empty | |
| + | plamid was transformed to the strain containing BBa_T9002, and a | ||
| + | Ampicillin resistance marker-containing empty vector was transformed | ||
| + | into the low copy variant, both resulting in a double transformation. | ||
| + | |||
| + | <br clear=all> | ||
===Method=== | ===Method=== | ||
Revision as of 03:37, 30 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
Copy number of Receiver Plasmid
Design
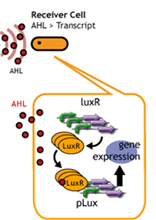
In this section, we intended to create time-delay by altering the
amount of luxR protein per cell.(*1) Since the amount of LuxR that
receives AHL differs, we hypothesized that the time required to reach
the threshold AHL concentration will differ.
To do this, we used different copy number plasmids of the Receivers. (*1)
By altering the copy number of plasmids in the receiver cells, the
time required for the response is altered.
If the copy number is altered, the amount of LuxR produced by the
receivers will also change.
The amount of LuxR that receives AHL will change, so the time required
for AHL to reach a threshold level will be altered.
Experiment
We used a pMB1 ori high copy vector containing BBa_T9002 and a low copy vector containing BBa_T9002 with a P15A as ori.
We determined the effects of altering the plasmid copy number by comparing the above situations.
Experiments used the following genetic circuits:
|
|
| **[http://partsregistry.org/Part:BBa_S03623 BBa_S03623 (AHL autoinucer)] |
|
|
In addition, a Chloramphenicol resistance marker-containing empty
plamid was transformed to the strain containing BBa_T9002, and a
Ampicillin resistance marker-containing empty vector was transformed
into the low copy variant, both resulting in a double transformation.
Method
- Transformed sender(Ptet-LuxI), high copy receiver(Ptet-LuxR-Plux-GFP-pMB1) and Medium copy receiver(Ptet-LuxR-Plux-GFP-p15A) respectively into E coli strains(JW1908).
- Inoculated them independently in liquid media. Incubated at 37°C 12h.
- Inoculated again in Fresh liquid media upto about OD600=2 at 37°C
- Washed sender and receivers.
- Mixed them. (Sender:Receiver=1000μL:1000μL)
- Incubated at 30°C.
- Measured intensity of green fluorescence at regular time intervals.(Fluoroskan AscentR FL&Fluoroskan AscentR Thermo ELECTRON CORPORATION)
Result & Discussion
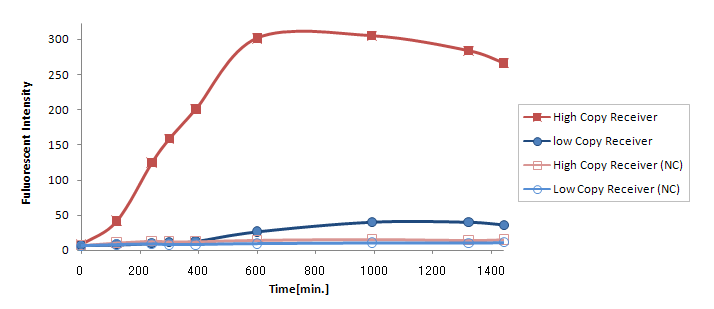
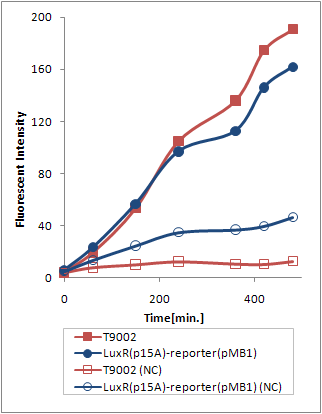
(Ptet-LuxR-plux-GFP-pMB1)の発現量に比べて(Ptet-LuxR-pLux-GFP-p15A)の発現量は大きく減少してしまった。(Fig.3.2)
この理由は
- GFPがローコピーのベクターに乗っていたので発現量が落ちた。
- LuxRの合成量が少なすぎる。
が考えられるが、(Ptet-luxR-p15A + plux-GFP-pMB1)と(Ptet-LuxR-pLux-GFP-pMB1)ではまったく同じTrasnfor Carveの軌跡を描いた。(Fig.3.3) このことより、p15Aのベクターに乗ったLuxRの発現量は、ハイコピーのベクターに乗ったpLuxを活性化するのに十分な量であると考えられる。 したがって理由1が有力である。
| Home | The Team | The Project | Parts Submitted to the Registry | Notebook |
|---|
 "
"