Team:HKUSTers/Modelling
From 2008.igem.org
(→Simulation Demo) |
(→Simulation Demo) |
||
| Line 136: | Line 136: | ||
[[Image:HKUSTers_simulation.zip|SimulationDemo]] | [[Image:HKUSTers_simulation.zip|SimulationDemo]] | ||
| + | ====User Interactive Simulation==== | ||
The upper part is User interactive: you can assign values to the main parameters, which are explained in the following sections. | The upper part is User interactive: you can assign values to the main parameters, which are explained in the following sections. | ||
| Line 147: | Line 148: | ||
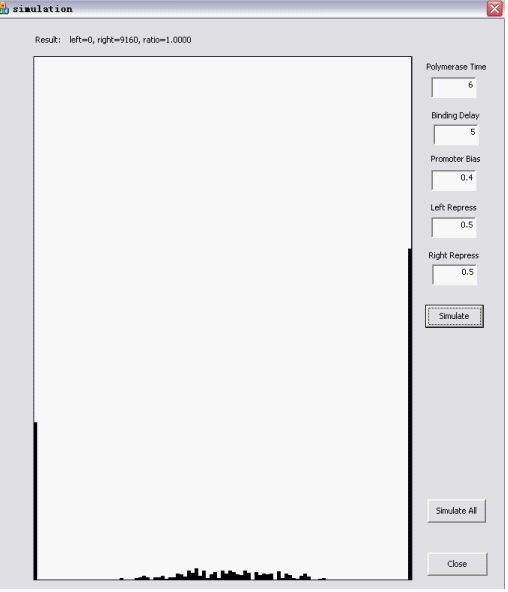
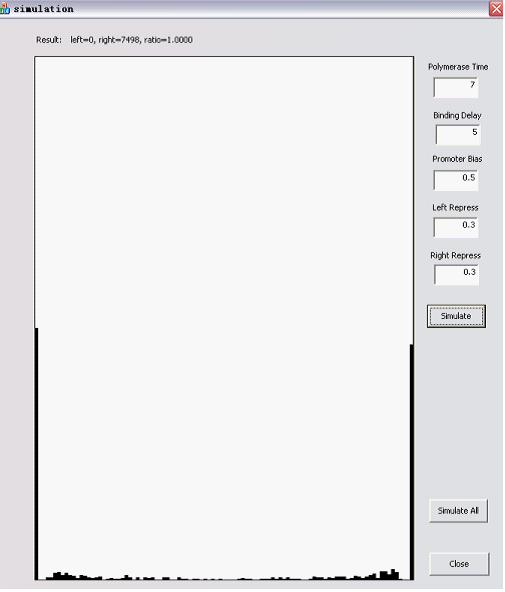
Here are two snapshots. | Here are two snapshots. | ||
| - | |||
| - | [[Image: | + | [[Image:HKsim1.png|450px|left]] |
| + | [[Image:HKsim2.png|450px|right]] | ||
| + | |||
| + | |||
| + | |||
| + | ===="Simulate All": global output with different parameter values==== | ||
If you click the "Simulate All" Button, an "output.txt" file would be generated that piles up key-information of all simulations from a predefined parameter range. | If you click the "Simulate All" Button, an "output.txt" file would be generated that piles up key-information of all simulations from a predefined parameter range. | ||
Revision as of 04:52, 30 October 2008
| Home | The Team | The Project | Parts | Modelling | Notebook | Gallery |
Simulation Demo
Please download and unzip this executable file, it's fun to play with the parameters!
User Interactive Simulation
The upper part is User interactive: you can assign values to the main parameters, which are explained in the following sections.
The Randomizer Performance (200 cells, each 100 Polymerase-Hit events, as elaborated in the following sections)is presented as a histogram:
- horizontal axis is r =(# of green proteins)/(# of green proteins + # of red proteins), ranging from 0 to 1 with 0.05 interval, r is N/A if total # is zero);
- vertical axis shows the summation of (number of) cells whose r value falls into the corresponding interval. The number of red/green proteins in individual cells are shown "real-time" to the program running at the upper left corner.
Here are two snapshots.
"Simulate All": global output with different parameter values
If you click the "Simulate All" Button, an "output.txt" file would be generated that piles up key-information of all simulations from a predefined parameter range.
Brief Description
- our design: based on random diffusion, signal amplified by reciprocal negative feedback loop
- simulation shows: when requirements for a critical parameter is met, the Randomizer is feasible and robust to other molecular fluctuations.
Assumptions (selected)
- T7 Polymerase concentration is low and stable over time
- Protein degradation is insignificant in the time scale of fate-decision (the first few mRNA transcription events has a preponderant effect)
- Cell devision is inhibited by a DNA-replication inhibitor, so the volume of the Cell remains constant (e.g. "total number" represents concentration).
- The expression of the Reporters of the two arms of the switch (GFP and RFP, respectively) is ideally coupled to the expression of the repressors at one-to-one ratio.
List of the Main Parameters
Description please see in the following section.
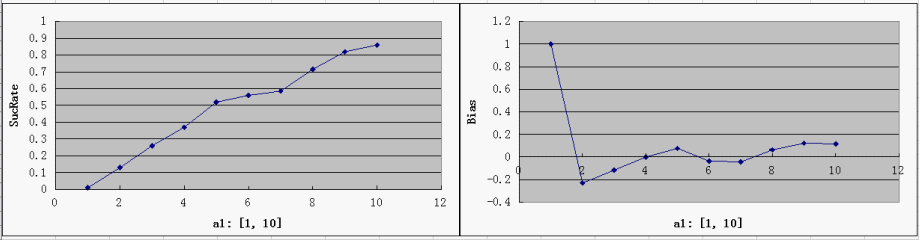
- a1 - time
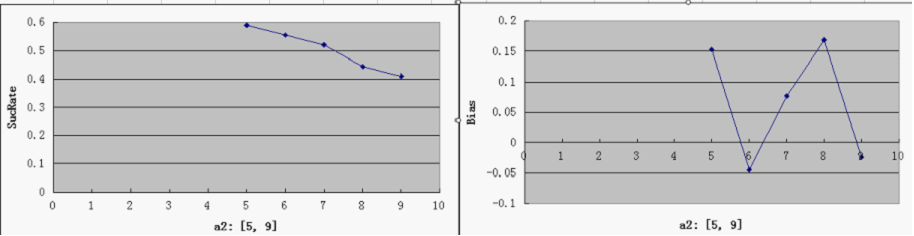
- a2 - time
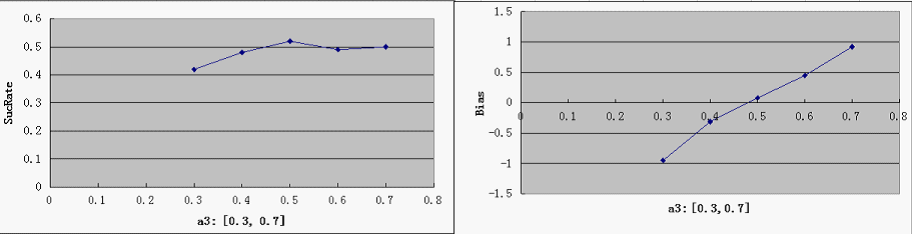
- a3 - probability of promoter bias
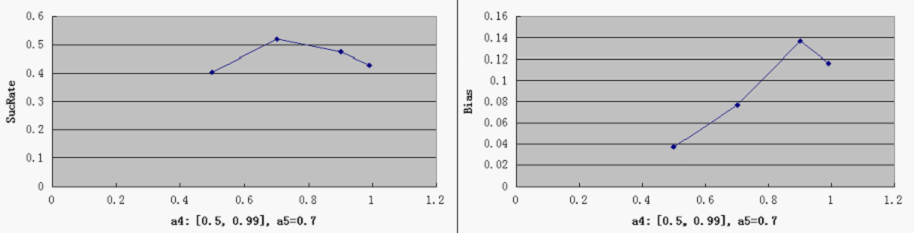
- a4 - probability of RepLeft blockage
- a5 - probability of RepRight blockage
(..under construction..)
Flow of Events
- One T7 RNA Polymerase binds to the bidirectional promotors (one "Pol-Hit"), with a probability of a3 binding to the LeftPromoter, then pauses (abortive transcription and conformational change). The time interval between 2 Pol-Hits is described by the variable "Hit-interval". This parameter fluctuates above a minimum of a1 seconds.
- Polymerase proceed into the elongation state but may be blocked by a repressor sitting at the repressor binding site. i.e. at Decision Timepoint, there is a certain probability that the event is terminated at this step.
- If the event is not blocked, transcription is followed by translation, and the nascent repressor protein folds, diffuses and may bind to its repressor binding site (on the other arm of the switch). In one Pol-Hit event, we define a variable "bindingDelay" as the time between the Decision Timepoint and timepoint where the first protein transcribed from this mRNA is functionally available. This variable fluctuates above a minimum of a2 seconds.
- We assume that the average life-time of this mRNA is around one minute, so in our model, after the first repressor protein is readily diffusing in the cell, a "burst" of proteins (number of molecules fluctuating between 10 and 20) are subsequently generated from this mRNA at an interval of 2 seconds.
- The repression at the repressor binding site is concentration dependent, which we simulate with
Probability of Effective Blockage = 1-(1-Eff)^n(t),
where Eff reflects the inherent efficiency of the repressor (assign parameters Eff = a4 for the RepLeft and Eff = a5 for the RepRight, Eff ∈(0,1)), and n(t) is the total number of available repressors at timepoint t. - This Loop of a single Pol-Hit Event ends here. As mentioned above, the next Pol-Hit occurs after a time-interval described by the variable "Hit-interval"(=a1+fluctuation).
- For each cell, we trace 100 successive Pol-Hit Events. 200 cells in total are sampled.
Simplified Qualitative Analysis & Discussion
Evaluation of Randomizer Performance
Two Criteria:
1. Our "Dice" - each cell - should deliver the binary output by displaying distinct red or green colour, not something yellowish... for each cell, Define an output as distinct if
1) (# of green proteins + # of red proteins) > 500 (demand sufficient strength of output) AND
2) r<0.05 or r>0.95, with r =(# of green proteins)/(# of green proteins + # of red proteins), corresponding to distinct red and distinct green, respectively for the total pool of cells, Define SuccessRate = (# of distinct cells)/(total # of cells)
2. The "Dice" should not be biased: Define Bias = (# of distinct green cells - # of distinct red cells)/ (# of distinct cells)
Behind the "Simulate All" Button...
- Initial (default) parameters, in 5 nested loops:
a1 = 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 a2 = 5, 6, 7, 8, 9 a3 = 0.3, 0.4, 0.5, 0.6, 0.7 a4 = 0.5, 0.7, 0.9, 0.99 a5 = 0.5, 0.7, 0.9, 0.99
- Output (for 100 Pol-Hits *200 cells):
SuccessRate Bias
Single Variable Plots
- For each parameter assigned as independent variable, keep the other variables at a median value (showed in italic)
- possible range for Y-axis and ideal value(good "dice" behavior):
SucRate: [0, 1], ideal: 1 Bias: [-1, 1], N/A if # of protein is 0, ideal: 0
With the aid of more sampling and plotting(unpresented data), we can preliminary conclude that
- the parameter a1 is the most critical one, since the Randomizer Performance heavily depends on a1 and strongly favors a larger a1. That focuses our attention on the control of T7 Polymerase during the design: a consistent low level of T7 Polymerases is desired, but too low a concentration would slow down the whole process of Random number generation, which is ineffective at best, and may even bring back adverse factors like protein degradation.
- predictably, Bias is clearly affected by a3, but it is not too sensitive - our symmetric bidirectional promoter design shall be robust enough.
- Compared to a3, other parameters do not by themselves have a decisive effect on good overall performance, at least within the medium range of general parameters settings. But noticeably, when the repressor efficiency is too high, there is a tendency for the switch to be "locked" in both directions - meaning that the total number of proteins is consistently very low (hence low SucRate), thus impairing the robustness of the system.
Under Construction
All parameters are approximate assignments, their estimatation based on biochemical data that we obtained from literature. Detailed Disscussion and partial justification of our choice of parameter values as well as model-building, along with the full list of references, is not documented in soft-copy yet.
the cryptic Code behind...
The simulation program written by Jiahui, in C++, please unzip after download:
File:SimulationDlg.zip "
"