Team:University of Alberta/Plastic Project:The University of Alberta
From 2008.igem.org
| Line 1: | Line 1: | ||
| - | + | ||
Revision as of 16:59, 2 July 2008
"How do I love thee, plastic? Let me count the ways. I wake up and glance at my plastic digital cable box to check the time. I go to the bathroom to use my plastic toothbrush, shaking a bit of my “nontoxic” tooth powder from a plastic bottle. I fill the plastic container of my Waterpik with mouthwash from another plastic bottle. I step into the shower—my lacy white curtain is protected by a plastic liner, and my chlorine-free shower water comes to me through a plastic-encased filter.Ah, but in the kitchen I am a bit freer of you, plastic. When I learned that my plastic bowls, dishes, and containers could leach harmful chemicals—especially the ones with that sneaky, practically invisible little recycling triangle embossed with the number 7—I bought Pyrex. My soft-boiled eggs are served up in a Pyrex glass dish. A moment of rebellion against thee, plastic! But the microwave in which I heat water for tea is made of plastic as well as metal. And the refrigerator shelf on which I store my eggs is plastic. The coaster on my desk, on which I place my steaming, oh-so-healthy green tea, is plastic. The 22-inch liquid-crystal computer monitor that seems to be the fulcrum of my entire existence is made of plastic. My keyboard, my mouse, my computer speakers, the CD cases for my music collection, my polycarbonate reading glasses, the remote control for my stereo, my telephone—all plastic. The sun shines through my window onto a riot of green plants in … plastic pots. (I could switch to ceramic, but it’s so heavy and hard to heft when I want to water them.) And I’ve been up for only an hour."
[http://discovermagazine.com/2008/may/18-think-you-can-live-without-plastic/(by Jill Neimark in Discovery Magazine 04.18.2008]
See also the article that inspired our project at [http://discovermagazine.com/2008/may/18-the-dirty-truth-about-plastic | Link]
The Project
The aim of our project is to create a biosensor that will detect BPA and other estrogenic compounds in the environment. BPA is a toxin that has been shown to leech from certain types of plastic. Studies have shown this chemical to have detremental effects in animal studies and is very likely to be harmful to humans as well. Having a biosensor that can detect soil and water contaminated with BPA would be a useful tool indeed.
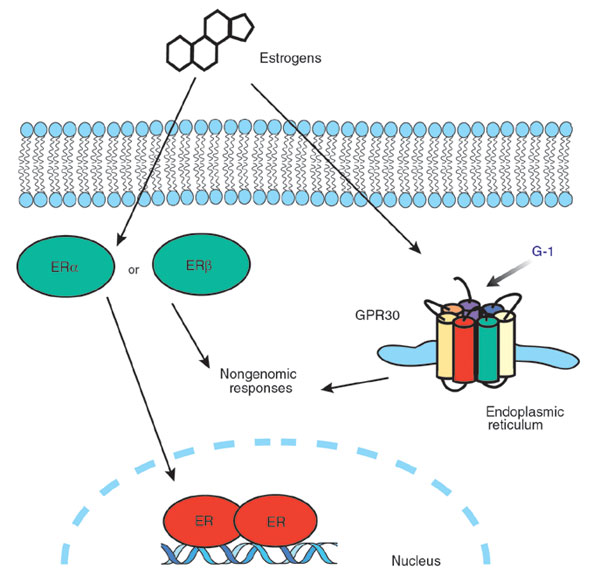
Our project relies on the mechanism by which BPA affects cells. BPA is an estrogenic compound; i.e. it has the ability to mimic estrogen. When taken into the cell, it can bind to the cytoplasmic estrogen receptors, and activate genes related to estrogen, tricking cells into thinking they are in an environment containing estrogen when they are not.
Our biosensor will take advantage of the estrogen receptor (see diagram to the right)
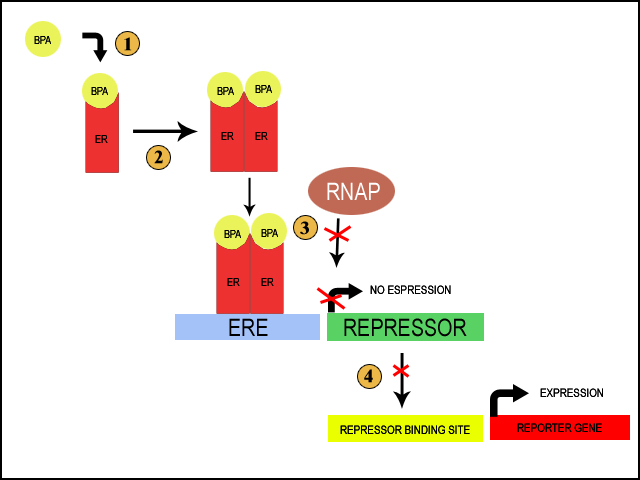
We aim to move the human estrogen receptor (hER) into E.Coli, which will activate a reporter system in the presence of BPA. When BPA is present in the environment, it difuses through the cell membrane and binds to free cytoplasmic estrogen receptor (1). This causes the estrogen receptors to dimerize (2). Dimerization of the estrogen receptors allows for the receptors to beind to the estrogen responsive element (ERE); this is a small sequence of DNA that the receptors can specifically bind once they are dimerized (3). The ERE is placed immeadetly upstream of the promotor for a repressor. The binding of the reseptor should then block the binding of RNA Polymerase and prevent the transcription of the repressor. Under normal circumstances, the repressor would itself prevent the transcription of a reporter gene (Purple Russian/Blue Ox). Thus, when BPA is in the environment, the repressor is repressed and the reporter gene is actively transcribed, giving us a quick visual way to determine the presence of BPA (4).
Next, plan on intorducing BPA degrading genes from Sphingomonas bisphenolicum to aid in the remediation of BPA contamination. Ultimately, we aim to put this biosensor/degradation system into plants - More on this later.
Info
- bisphenol degrading gene from the Sphingomonas bisphenolicum strain AO1
[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=171363657 Link]
Proposed Work Timeline: A Work In Progress
Proposed Timeline: Plastic Project
- June 12: Group meeting, submission of Bio-brick parts for BPA system
- June 13: Final review of Bio-bricks to be submitted
o Check sequences to make sure that there are not any unwanted restriction sites or stop codons.
o Consider optimizing the sequences… I don’t think that they need to be optimized for high expression, but if there are any truly inefficient codons, we should switch those.
WEEK 1 (June 16-20):
- Butanerd work
- Gardening
- Design and order all the primers that we will need for sequencing our biobricks, bearing in mind that some of our bricks are too long to be properly sequenced just by Vf and Vr primers and will need internal sequencing primers.
- Chose a plasmid to be our operon’s backbone. We’ll need to consider what promoter it carries and the copy number.
WEEK 2 (June 23-27):
- Arrival of Biobricks
- If biobricks are made in a plasmid, transform the plasmid into E. coli, and freeze down a few cultures at -80C.
- If they do not come in a plasmid, but as linear DNA, ligate them into a plasmid and freeze down samples of that culture.
- Sequence the biobricks from the plasmid using our sequencing primers
- Test the prefix and suffix restriction sites on the biobricks plasmid. You should be able to get the bricks out of the plasmid with double digests (E/X+P/S) or a single digest with NotI.
- Make concentrated gel purifications of all the individual biobricks after they have been digested with EcoRI and SpeI.
- Make concentrated gel purifications of the backbone plasmid after it is digested with EcoRI and XbaI
- Gardening
WEEK 3 (June 30-July 4):
- We will be making our operon with 5 genes in sequence ERα, TetR, Reporter Gene (Purple Russian or Blue Ox), BisdA, and BisdB. To make the operon all in one plasmid, we will use the rolling assembly strategy outlined on the igem page. (note: I think that we should try and make a few different operons in addition to the complete one, like one without the degradation genes and one without the visual reporters)
- Ligate the gel purified TetR fragment into the gel purified backbone plasmid (From ligation to a confirmation digest being run on a gel, this should take 3 days, maybe 4)
- At the same time but in a separate reaction, ligate the gel purified BisdB into the gel purified backbone plasmid
- At the same time but in a separate reaction, ligate the gel purified Purple Russian into the gel purified backbone plasmid
- At the same time but in a separate reaction, ligate the gel purified Blue Ox into the gel purified backbone plasmid
- At the end of the week we should have plasmids containing TetR, Purple Russian, Blue Ox and BisdB, but don’t bother sequencing them at this point.
- Digest the TetR plasmid with EcoRI and XbaI and gel purify.
- Digest the BisdB plasmid with EcoRI and XbaI and gel purify.
- Digest the Purple Russian and Blue Ox plasmids with EcoRI and SpeI and gel purify.
WEEK 4 (July 7-11):
- Grow up cultures for the E.coli strains containing the successful TetR, Purple Russian, Blue Ox and BisdB plasmids from the previous week.
- Look at the Purple Russian and Blue Ox cultures under a fluorescent microscope (using untransformed E. coli as a control)
- Purify protein from the cultures and do a Western Blot to test for expression of TetR and BisdB (both should express in their current configuration) and to get an idea about how they migrate on the gel.
- Ligate the E+X digested TetR plasmid from week 3 with the E+S digested ERα fragment from week 2 and transform into E. coli. Digest to confirm transformation.
- Ligate the E+X digested BisdB plasmid from week 3 with the E+S digested BisdA fragment from week 2 and transform into E. coli. Digest to confirm transformation.
WEEK 5 (July 14-18):
- Grow up successful plasmids from week 4 and test for protein expression using a Western Blot. There should be two bands for each of the new strains. If we have estrogen and BPA we should test to see if the ERα+TetR plasmid can have TetR expression turned off by either estrogen or BPA.
- Digest the ERα+TetR plasmid from week 4 with EcoRI and SpeI and gel purify the insert.
- Digest the BisdA+BisdB plasmid from week 4 with EcoRI and XbaI and gel purify.
- Ligate the gel purified EcoRI and XbaI digested BisdA+BisdB plasmid with the EcoRI and SpeI gel purified ERα+TetR fragment (transform/check).
- Ligate the gel purified EcoRI and XbaI digested BisdA+BisdB plasmid with the EcoRI and SpeI gel purified Purple Russian fragment (transform/check).
- Ligate the gel purified EcoRI and XbaI digested BisdA+BisdB plasmid with the EcoRI and SpeI gel purified Blue Ox fragment (transform/check).
- Ligate the gel purified EcoRI and XbaI digested Purple Russian plasmid with the EcoRI and SpeI gel purified ERα+TetR fragment (transform/check).
- Ligate the gel purified EcoRI and XbaI digested Blue Ox plasmid with the EcoRI and SpeI gel purified ERα+TetR fragment (transform/check).
- At this point we should have a working estrogen sensitive BisdA/B plasmid as well as estrogen sensitive plasmids for both optical reporters.
WEEK 6 (July 21-25):
- This is where we start biochemical tests. We need to figure out how we want to do this. Basically we need to test how well the different strains deal with the introduction of BPA into the system. Do they light up/break down the BPA? What happens to the protein levels?
- Time to make the last plasmids with the complete operons.
- Digest the ERα+TetR+Purple Russian plasmid with EcoRI and SpeI and gel purify the insert and ligate it into the previously EcoRI and XbaI digested and gel purified BisdA+BisdB plasmid (transform/confirm).
- Digest the ERα+TetR+Blue Ox plasmid with EcoRI and SpeI and gel purify the insert and ligate it into the previously EcoRI and XbaI digested and gel purified BisdA+BisdB plasmid (transform/confirm).
WEEK 7 (July 28-Aug. 1):
WEEK 8 (Aug. 4-8):
WEEK 9 (Aug. 11-15):
WEEK 10 (Aug. 18-22):
WEEK 11 (Aug: 25-29):
 "
"