Team:Hawaii/PCR Amplification of pRL1383a
From 2008.igem.org
(Difference between revisions)
m (→Results) |
m (→Results) |
||
| Line 66: | Line 66: | ||
* <br> | * <br> | ||
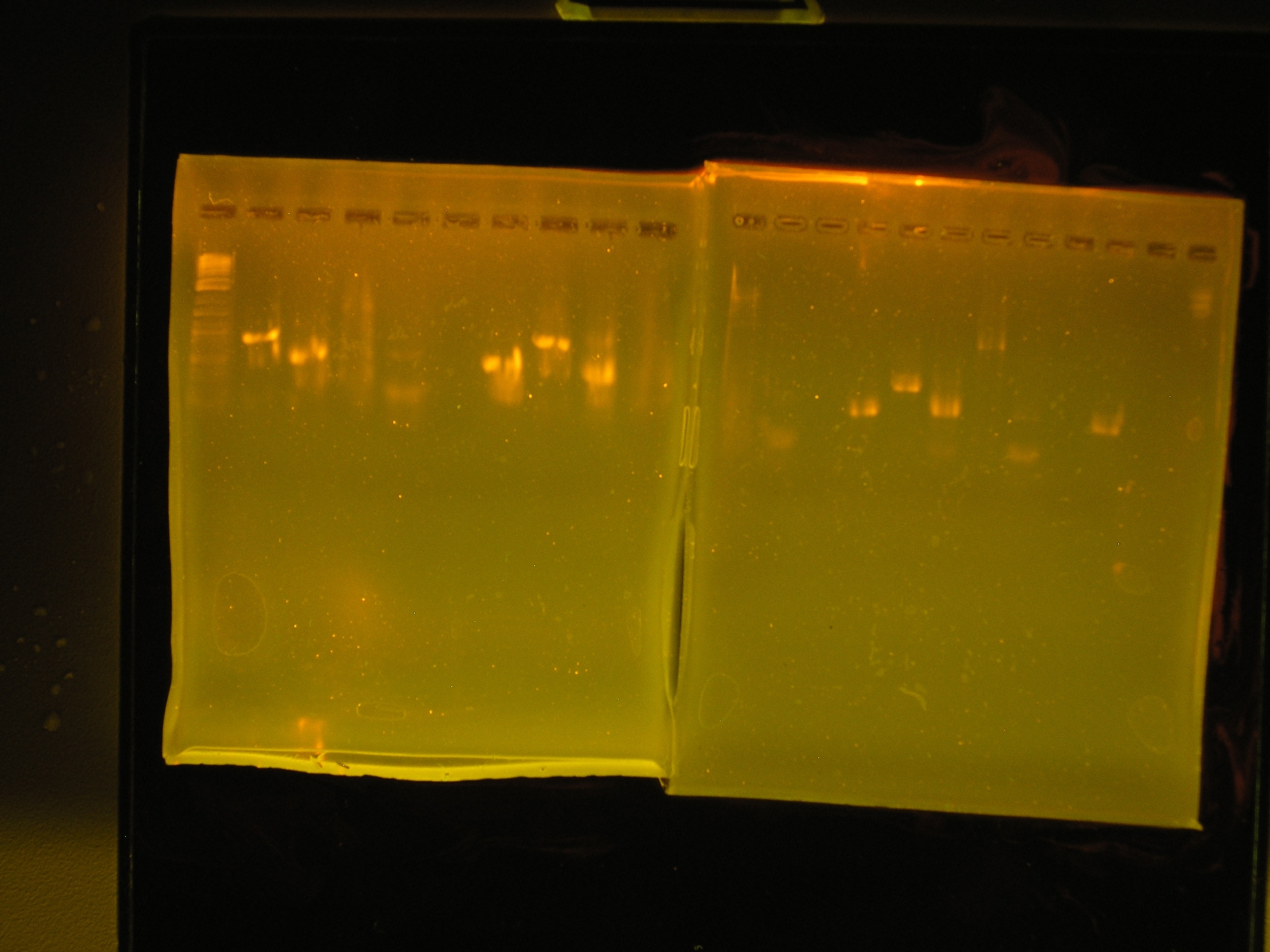
| - | + | [[Image:PCR_prl1383a.JPG|right|thumb|300px|PCR Gel 1 (left) and PCR Gel 2 (right).]] | |
{| border="1" | {| border="1" | ||
|+ '''PCR Gel 1''' | |+ '''PCR Gel 1''' | ||
| - | !width=" | + | !width="100"|Lane |
| - | !width=" | + | !width="100"|Contents |
| - | !width=" | + | !width="100"|Description |
| - | !width=" | + | !width="100"|Predicted annealingT°C,5 cycles |
| - | !width=" | + | !width="100"|Predicted annealing T°C,30 cycles |
|- | |- | ||
|1 | |1 | ||
| Line 136: | Line 136: | ||
|} | |} | ||
| - | |||
{| border="1" | {| border="1" | ||
|+ '''PCR Gel 2''' | |+ '''PCR Gel 2''' | ||
| - | !width=" | + | !width="100"|Lane |
| - | !width=" | + | !width="100"|Contents |
| - | !width=" | + | !width="100"|Description |
| - | !width=" | + | !width="100"|Predicted annealing T°C,5 cycles |
| - | !width=" | + | !width="100"|Predicted annealing T°C,30 cycles |
|- | |- | ||
|1 | |1 | ||
Revision as of 00:35, 4 July 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Contents |
PCR Amplification of pRL1383a
- Several regions of pRL1383a will be amplified with BioBrick based primers. These components will be used later in the construction of a pRL1383a BioBrick based vector. These parts include the aadA region of the omega interposon, the origin of vegetative replication (oriV), the mobilization proteins, and the replication proteins.
Methods
Materials Per Reaction Added in order:
- 3.5uL nanopure water
- 1uL 10uM forward/reverse primers
- 1uL pRL1383a as template (1:10 dilution of [Team:Hawaii/Header: Large-Scale Plasmid Prep from E. coli| large-scale plasmid prep].
- 5uL Taq polymerase: Accusure(hot start, 68°C) and Red Taq
Note: the taq and water can be combined and aliquoted together as long as reaction is kept at 4°C.
- cold blocks
- pcr reaction tubes, with tops
- pipette and tips
| Duration of Time | T° 5 cycles | T° 30 cycles | Purpose |
|---|---|---|---|
| 10 minutes | 95°C | 95°C | heat activate taq |
| 30 seconds | 95°C | 95°C | denaturation |
| 30 seconds | 48.1°C, 53.4°C, 57.9°C | 59°C, 60.6°C, 61.9°C | annealing |
| 5 minutes | 68°C | 68°C | extension (1.5 minutes per kb, go with longest) |
| 10 minutes | 68°C | 68°C | finishing the extension |
| infinity | 4°C | 4°C | (until you remove product) |
Materials for the gel
- 1% agarose gel
- 10ug/mL Ethidium Bromide
- running apparatus
- gel running conditions: 95V for ~1 hour
Results
| Lane | Contents | Description | Predicted annealingT°C,5 cycles | Predicted annealing T°C,30 cycles |
|---|---|---|---|---|
| 1 | Tri-dye 10kb ladder | ok | ||
| 2 | omega (aadA)#1 | (+), ~1kb | 54.4,50.4 | 60.7,69.5 |
| 3 | oriV #1 | (+), ~0.5kb | 51.3, 53 | 60.9, 71.2 |
| 4 | mob #1 | smear | 53.5, 48 | 62, 69.8 |
| 5 | rep #1 | smear, low kb | 53.9, 57.2 | 59.1, 73.5 |
| 6 | (-)control | all clear | ||
| 7 | (+) control | bright band, ~0.5kb | ||
| 8 | omega #2 | (+), ~1kb | 54.4,50.4 | 60.7,69.5 |
| 9 | oriV #2 | (+) ~0.5kb | 51.3, 53 | 60.9, 71.2 |
| 10 | mob #2 | nothing, slight smear | 53.5, 48 | 62, 69.8 |
| Lane | Contents | Description | Predicted annealing T°C,5 cycles | Predicted annealing T°C,30 cycles |
|---|---|---|---|---|
| 1 | Tri-dye 10kb ladder | no ladder | ||
| 2 | rep#2 | light smear | 53.9, 57.2 | 59.1, 73.5 |
| 3 | (-)control | all clear | ||
| 4 | (+)control | bright band ?0.5kb | ||
| 5 | omega #3 | bright band, ?1kb | 54.4,50.4 | 60.7,69.5 |
| 6 | oriV #3 | bright band, 0.5kb? | 51.3, 53 | 60.9, 71.2 |
| 7 | mob | light band, some smear, 3kb? | 53.5, 48 | 62, 69.8 |
| 8 | rep #3 | band in low kb | 53.9, 57.2 | 59.1, 73.5 |
| 9 | (-) control | all clear | ||
| 10 | (+) control | bright band, 0.5kb? |
Discussion
- What was learned and how to do future experiments differently.
Insanity is doing the same thing over and over again and expecting different results. - Albert Einstein
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"