Team:NYMU-Taipei/Project/Attachment
From 2008.igem.org
(Difference between revisions)
Blackrabbit (Talk | contribs) |
Blackrabbit (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Header}} | {{:Team:NYMU-Taipei/Header}} | ||
== Motivation == | == Motivation == | ||
| - | * | + | * Using gene-transformed bacteria in medical way is currently a hot biology and medicine topic, so we wonder about one thing: is there any way to boost the medical ability of bacteria? It's well known that many species of bacteria have the ability to attach to human tissue or epithelial cells. If we can control the attaching/detaching mechanism of these gene-transformed bacteria, we can enhance the bacteria ability by making a more direct interaction with a more specific targets, thereby make the whole system work more efficiently. |
| - | == | + | == Goals == |
| - | *When genetic ''E.coli'' senses the alternation of pH in intestine, the adhesion mechanism will be turned on in order to '''<font color="red">enhance</font>''' the ability of attachment. | + | *Design a genetic ''E.coli'' to remove waste through the small intestine epithelium. |
| - | *The enhanced adhesion will | + | *When our genetic ''E.coli'' senses the alternation of pH in the small intestine, the adhesion mechanism will be turned on in order to '''<font color="red">enhance</font>''' the ability of attachment. |
| - | *To cooperate with the "[[Team:NYMU-Taipei/Project/Time Regulation|Time Regulation]]" | + | *The enhanced adhesion will give our genetic ''E.coli'' more time to remove the wastes removed by the [[Team:NYMU-Taipei/Project/Waste Removal|Waste Removal]] subgroups. |
| - | + | *To cooperate with the "[[Team:NYMU-Taipei/Project/Time Regulation|Time Regulation]]" subgroup to allow our genetic ''E.coli'' to detach from intestinal epithelia cells after a specified amount of time. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==What to Use to Attach to the Small Intestine== | ||
| + | *We begin with three candidates: '''<font color="red">FimH</font>''', '''<font color="red">FliC</font>''' and '''<font color="red">intimin</font>''', and compare them below. | ||
| + | ====<font color="blue">FimH</font>==== | ||
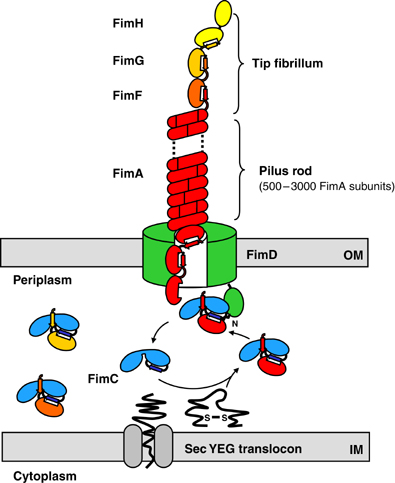
| + | *The FimH protein is the receptor recognizing element of type 1 fimbriae | ||
{| | {| | ||
| | | | ||
[[Image:NYMU_7600693f1.jpg]] | [[Image:NYMU_7600693f1.jpg]] | ||
| | | | ||
| - | + | ||
*Fimbriae | *Fimbriae | ||
| - | **Function: attachment of bacteria to its host as in case of | + | **Function: attachment of bacteria to its host as in the case of bacteria ''Escherichia coli'', ''salmonella typhimurium'', ''Nisseria gonorrhoea'', and ''bordella pertussis''. |
| - | **Size and number: Found many in number (up to 1000 per cell) having 3 to 25 nm | + | **Size and number: Found many in number (up to 1000 per cell), each having a diameter of between 3 to 25 nm, and each being 0.5 to 20 micrometers in length. |
* '''Type 1 fimbriae''' are surface organelles of ''Escherichia coli'' which mediate '''D-mannose-sensitive binding''' to different host surfaces. | * '''Type 1 fimbriae''' are surface organelles of ''Escherichia coli'' which mediate '''D-mannose-sensitive binding''' to different host surfaces. | ||
| - | * A single type 1 fimbria is a 7-nm-wide and »1- | + | * A single type 1 fimbria is a 7-nm-wide and »1-um-long surface polymer, the bulk of the structure is made up of »1000 copies of the major subunit protein, FimA, polymerized into a right-handed helical structure. |
* Small quantities of the minor components (FimF, FimG, and FimH) are also present. | * Small quantities of the minor components (FimF, FimG, and FimH) are also present. | ||
| - | * The '''FimH''' protein is the '''receptor recognizing element''' of type 1 fimbriae. | + | * The [[Team:NYMU-Taipei/Project/Attachment/FimH|'''FimH''']] protein is the '''receptor recognizing element''' of type 1 fimbriae. |
* Studies on ''Salmonella Typhimurium'' revealed that FimH adhesin is responsible for bacteria binding to HeLa, HEp-2 and mouse intestinal epithelial cells. | * Studies on ''Salmonella Typhimurium'' revealed that FimH adhesin is responsible for bacteria binding to HeLa, HEp-2 and mouse intestinal epithelial cells. | ||
| + | *Research indicats that bacteria with FimH-FocH hybrids still can act as normal bacteria, while the bacteria with ''fimH'' mutation loses its ability to attach to intestine. | ||
|- | |- | ||
|} | |} | ||
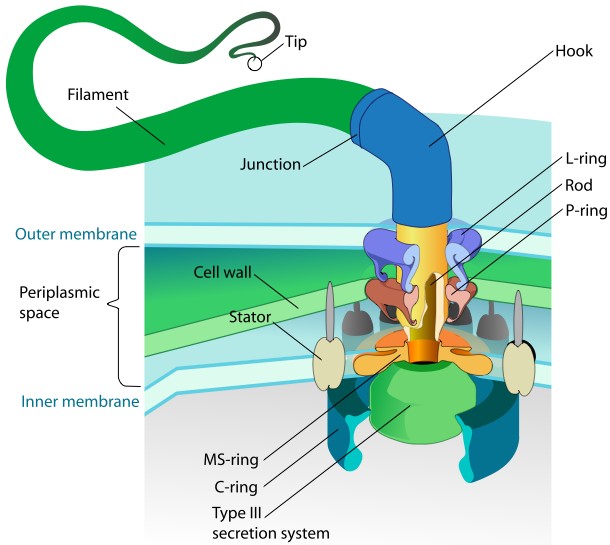
| - | == | + | ====<font color="blue">FliC</font>==== |
{| | {| | ||
| | | | ||
[[Image:NYMU_Flagellum.png]] | [[Image:NYMU_Flagellum.png]] | ||
| | | | ||
| - | *FliC is an antigen of E.coli | + | *FliC is an antigen of '''''E.coli'' flagellum'''. |
*It binds to '''mucin''' and ECM. | *It binds to '''mucin''' and ECM. | ||
| + | *We didn't choose it to use because there are four PstI cuttting sites on K12 ''fliC'' gene. | ||
| + | *However, it still can be one of our models. | ||
|- | |- | ||
|} | |} | ||
| - | == Circuit Design == | + | ====<font color="blue">Intimin</font>==== |
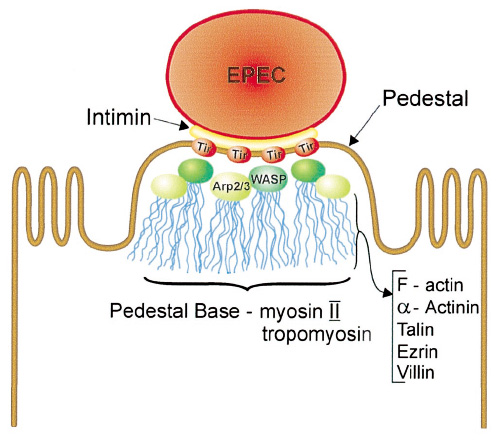
| - | * | + | {| |
| - | **Design | + | | |
| - | ** | + | [[Image:NYMU_PROCESS3.jpg]] |
| + | | | ||
| + | *Intimin is an adhesin which includes in a protein collection '''Esps(EPEC secreted proteins'''). | ||
| + | *It is coded in ''eae'' gene, which is a part of pathogenicity island termed '''the locus of enterocyte effacement''' '''(LEE).''' | ||
| + | *Intimin can binds to host cell either through [http://en.wikipedia.org/wiki/Type_three_secretion_system#Type_III_secretion_system_.28T3SS.29|'''type III secretion system''' ], or via cell receptor, '''integrin'''. | ||
| + | *Studies of intimin reaval that even intimin domain could cause unsual cytoskeleton rearrange ment, which could lead to A/E leison; Therefore, we didn't use it as model. | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | == How to Attach in Intestine == | ||
| + | *We enhance the attachment in intestine by enhancing the expression of the membrane form of '''<font color="red">FimH</font>'''. | ||
| + | [[Image:NYMU_Attachment_pic.jpg]] | ||
| + | *<font color="red">We can use the biobrick part!</font> | ||
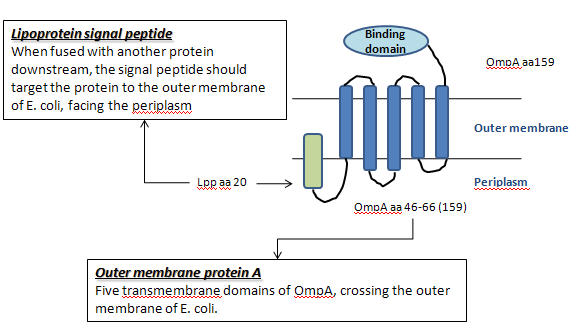
| + | **Lac-inducible generator of Lpp-OmpA(46-66)-Streptavidin wild-type+ His6 tag('''Part:BBa_J36848''')[http://partsregistry.org/Part:BBa_J36848] | ||
| + | ***This device contains a lac promoter and strong ribosome binding site for lac-inducible expression of the fusion protein of Lpp signal peptide, OmpA aa46-66, and streptavidin wild-type + His6 tag. This expression should display streptavidin on the cell surface of ''E. coli''. | ||
| + | ***By displace the lac promoter and streptavidin part with '''pH promoter''' and '''FimH binding domain''', we can induce the expression of FimH in an optimal pH, thus enhance the adhesion of genetic ''E.coli'' in intestine. | ||
| + | |||
| + | ==How to Detach in Intestine == | ||
| + | *[[image:NYMU_Attachment_protease-5.jpg]] | ||
| + | *[[image:NYMU_Attachment_cuttingsite-5.jpg]] | ||
| + | *We '''design a cutting site between FimH and trans-membrane domain''', than we tfansform a '''protease''' gene into our ''E.coli''. | ||
| + | *The protease gene can be regulated by the promoter of '''time regulation''', giving the expression of a secreted protease, which can cut the cutting site we design. | ||
| + | |||
| + | ==Circuit Design and Methods== | ||
| + | ====Construct==== | ||
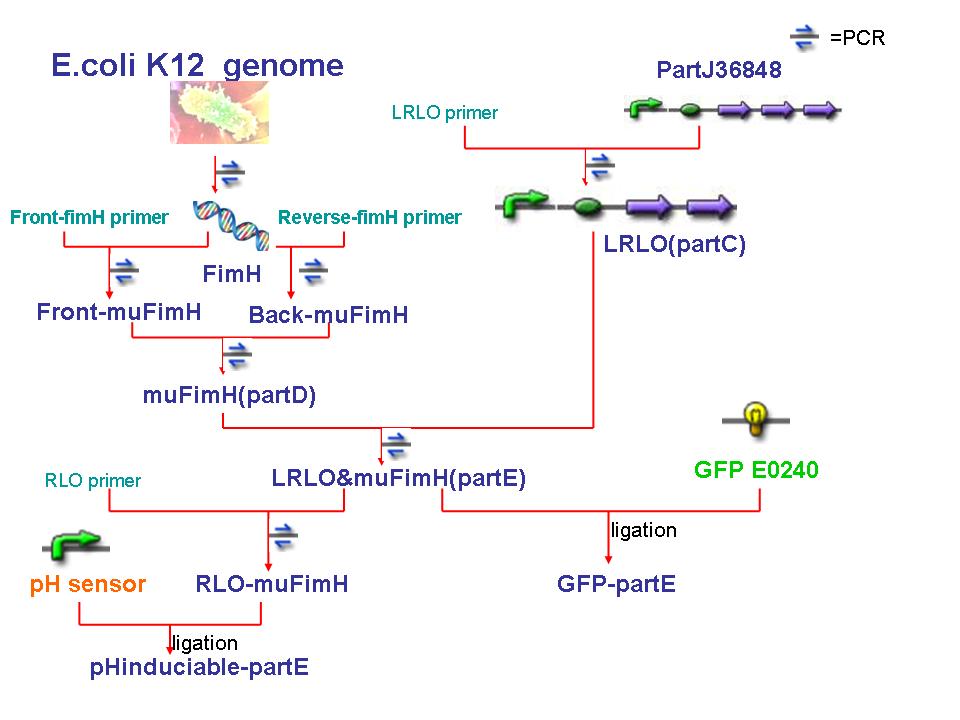
| + | [[image:NYMU NYMU-Attachment-constructnew.jpg]] | ||
| + | *We begin our work at gaining FimH and LRLO. | ||
| + | *FimH is from E.coli K12. We uses its genome as template and use PCR to gain FimH. | ||
| + | * As mentioned in "'''How to Attach in Intestine'''", there are five units in partJ36848 : a lacZ promoter, a ribosome binding site , Lpp aa 20, OmpA aa46-66 and streptavidin + His6 tag. Because we only need the first fout parts, so we use PCR to produce the sequence we need, LRLO or partC.(J36848 without streptavidin) | ||
| + | *<font color="blue">The reverse primer of LRLO is designed complemented to the front primer of muFimH.</font> | ||
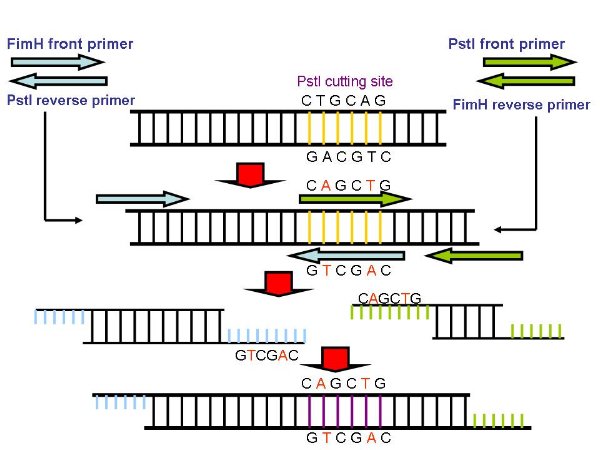
| + | *Because there is a PstI cutting site in FimH, so we use a PCR method to mutate the cutting site. We desigh two set of primers, primerA('''pA''') anr primerB('''pB'''). | ||
| + | [[image:NYMU Attachment-muFimH-5.jpg]] | ||
| + | **The front primer of pA and the reverse primer of pB is actually the primers we use to gain FimH, While pA reverse primer and pB front primer are designed to aneal to the two strand of PstI cutting site, while one of the base pair has been replaced. | ||
| + | **We use pA and pB to gained the front part of FimH as well as back part seperately. Because the reverse pA and front pB are complemented, so the two parts can be linked together by PCR, and since one of the basepair has been replaceed, so the cutting site will be mutated, thus we gain mutant FimH(muFimH, we also call it partD). | ||
| + | *After we gained muFimH and LRLO , we use PCR to link the two parts as the primer are complemented. Finally we get LRLO-muFimH sequence(partE), the main constructed sequence in our project. | ||
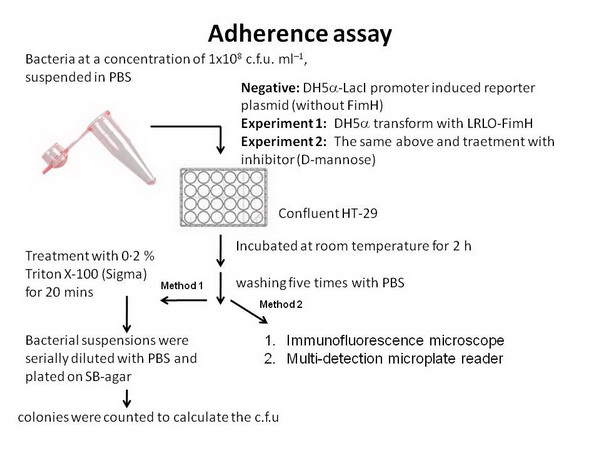
| + | *Before combining with other group's design, we need to test ours. Therefore we do two binding assay: | ||
| + | **One use GFP-partE to observe wether the ability of LRLO-FimH hybrids can attach to intestine. | ||
| + | **The other use pH sensor to replace lacZ promoter to see wether the protein expression could be regulate by pH. | ||
| + | |||
| + | ====Attachment Circuit Design==== | ||
| + | |||
| + | *'''WORKING PART''' | ||
| + | *[[Image:NYMU_Attachment_bindingcircuit-5.jpg]] | ||
| + | *'''TESTING PART''' | ||
| + | *[[Image:NYMU_Attachment_testingcircuit-5.jpg]] | ||
| + | |||
| + | ====Detachment Circuit Design==== | ||
| + | *'''PROTEASE PART''' | ||
| + | *[[image:NYMU Attachment protease-5.jpg]] | ||
| + | *Design protease cleavage site between OmpA and FimH | ||
| + | *[[image:NYMU Attachment cuttingsite-5.jpg]] | ||
| + | |||
| + | == Experiment Result == | ||
| + | *'''Protocal and Method Check out''' | ||
| + | ** Methods | ||
| + | **[[image:NYMU_attach_protocal.jpg]] | ||
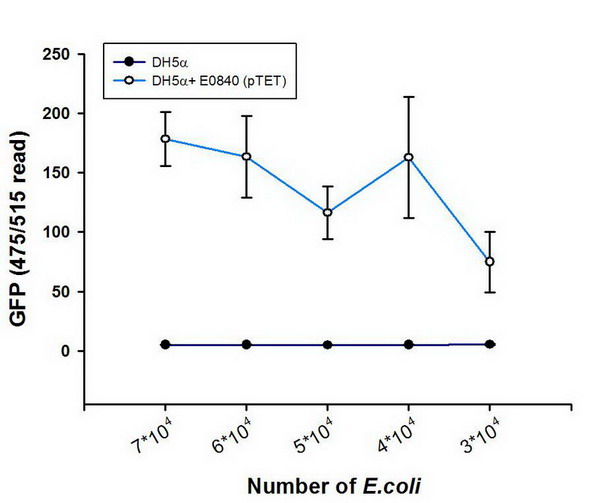
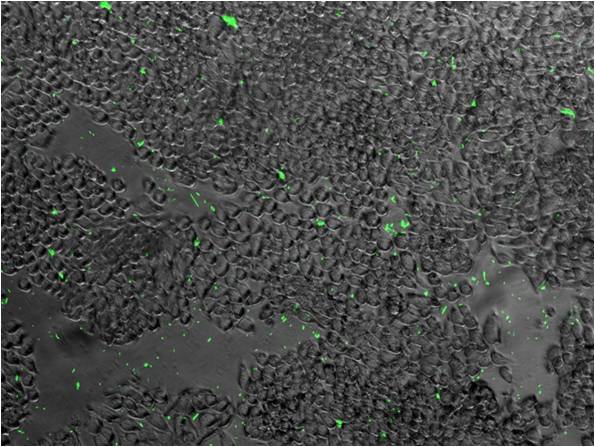
| + | ***'''Figure A. '''DH5a transformed with E0840 (pTET) reporter plasmid can be detected by multi-detection microplate reader. | ||
| + | ***[[image:NYMU_Figure.1.jpg]] | ||
| + | ***'''Figure B. ''' HT29 cells incubated with DH5a transformed with E0840 (pTET) '''GFP''' reporter plasmid. | ||
| + | ***[[image:NYMU_Figure.2.jpg]] | ||
== References == | == References == | ||
| - | *Functional characterization of the FimH adhesin from Salmonella enterica serovar Enteritidis[http://mic.sgmjournals.org/cgi/content/full/152/5/1337?view=long&pmid=16622051] | + | *<font color="blue">Functional characterization of the FimH adhesin from Salmonella enterica serovar Enteritidis</font>(D. Kisiela etc,2006)[http://mic.sgmjournals.org/cgi/content/full/152/5/1337?view=long&pmid=16622051] |
| - | *Combining sites of bacterial fimbriae [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6VS6-4PGPKS8-2&_user=1576506&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000053839&_version=1&_urlVersion=0&_userid=1576506&md5=33dfc6974f43b7ece79e0e7b90044ee7] | + | *<font color="blue">Combining sites of bacterial fimbriae</font>(H. D. Grevea, L. Wynsa, J. Bouckaerta,2007) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6VS6-4PGPKS8-2&_user=1576506&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000053839&_version=1&_urlVersion=0&_userid=1576506&md5=33dfc6974f43b7ece79e0e7b90044ee7] |
*Probing the receptor recognition site of the FimH adhesin by fimbriae- displayed FimH-FocH hybrids[http://mic.sgmjournals.org/cgi/reprint/144/7/1919?view=reprint&pmid=9695925] | *Probing the receptor recognition site of the FimH adhesin by fimbriae- displayed FimH-FocH hybrids[http://mic.sgmjournals.org/cgi/reprint/144/7/1919?view=reprint&pmid=9695925] | ||
| - | *Host Protein Binding and Adhesive Properties of H6 and H7 Flagella of Attaching and Effacing Escherichia coli[http://jb.asm.org/cgi/content/full/189/20/7426] | + | *<font color="blue">Host Protein Binding and Adhesive Properties of H6 and H7 Flagella of Attaching and Effacing Escherichia coli</font>(A. L. Erdem etc,2007)[http://jb.asm.org/cgi/content/full/189/20/7426] |
| - | *Sequence Diversity of the Escherichia coli H7 fliC Genes:Implication for a DNA-Based Typing Scheme forE. coli O157:H7[http://jcm.asm.org/cgi/reprint/38/5/1786.pdf] | + | *<font color="blue">Sequence Diversity of the Escherichia coli H7 fliC Genes:Implication for a DNA-Based Typing Scheme forE. coli O157:H7</font>(L. WANG etc,2007)[http://jcm.asm.org/cgi/reprint/38/5/1786.pdf] |
| + | *<font color="blue">Exploitation of host cells by enteropathogenic Escherichia coli</font>(B. A. Vallance and B. B. Finlay,2000)[http://cmgm.stanford.edu/micro/MI209/EPEC.pdf] | ||
| + | *<font color="blue">The Cell-binding Domain of Intimin from Enteropathogenic Escherichia coli Binds to b1 Integrins</font>(G. Dougan etc,1996)[http://www.jbc.org/cgi/reprint/271/34/20359] | ||
| + | *<font color="blue">Intimin from enteropathogenic Escherichia coli mediates remodelling of the eukaryotic cell surface</font>(A.D.Phillips etc,2000)[http://mic.sgmjournals.org/cgi/content/full/146/6/1333] | ||
| + | *<font color="blue">Intervening with Urinary Tract Infections Using Anti-Adhesives Based on the Crystal Structure of the FimH–Oligomannose-3 Complex</font>(A. Wellens etc,2008)[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2323111] | ||
| + | |||
{{:Team:NYMU-Taipei/Footer}} | {{:Team:NYMU-Taipei/Footer}} | ||
Revision as of 19:31, 29 October 2008
| Home | Project Overview: | pH Sensor | Attachment | Time Regulation | Waste Removal | Experiments and Parts | About Us |
Contents |
Motivation
- Using gene-transformed bacteria in medical way is currently a hot biology and medicine topic, so we wonder about one thing: is there any way to boost the medical ability of bacteria? It's well known that many species of bacteria have the ability to attach to human tissue or epithelial cells. If we can control the attaching/detaching mechanism of these gene-transformed bacteria, we can enhance the bacteria ability by making a more direct interaction with a more specific targets, thereby make the whole system work more efficiently.
Goals
- Design a genetic E.coli to remove waste through the small intestine epithelium.
- When our genetic E.coli senses the alternation of pH in the small intestine, the adhesion mechanism will be turned on in order to enhance the ability of attachment.
- The enhanced adhesion will give our genetic E.coli more time to remove the wastes removed by the Waste Removal subgroups.
- To cooperate with the "Time Regulation" subgroup to allow our genetic E.coli to detach from intestinal epithelia cells after a specified amount of time.
What to Use to Attach to the Small Intestine
- We begin with three candidates: FimH, FliC and intimin, and compare them below.
FimH
- The FimH protein is the receptor recognizing element of type 1 fimbriae
|
FliC
|
Intimin
|
How to Attach in Intestine
- We enhance the attachment in intestine by enhancing the expression of the membrane form of FimH.
- We can use the biobrick part!
- Lac-inducible generator of Lpp-OmpA(46-66)-Streptavidin wild-type+ His6 tag(Part:BBa_J36848)[http://partsregistry.org/Part:BBa_J36848]
- This device contains a lac promoter and strong ribosome binding site for lac-inducible expression of the fusion protein of Lpp signal peptide, OmpA aa46-66, and streptavidin wild-type + His6 tag. This expression should display streptavidin on the cell surface of E. coli.
- By displace the lac promoter and streptavidin part with pH promoter and FimH binding domain, we can induce the expression of FimH in an optimal pH, thus enhance the adhesion of genetic E.coli in intestine.
- Lac-inducible generator of Lpp-OmpA(46-66)-Streptavidin wild-type+ His6 tag(Part:BBa_J36848)[http://partsregistry.org/Part:BBa_J36848]
How to Detach in Intestine


- We design a cutting site between FimH and trans-membrane domain, than we tfansform a protease gene into our E.coli.
- The protease gene can be regulated by the promoter of time regulation, giving the expression of a secreted protease, which can cut the cutting site we design.
Circuit Design and Methods
Construct
- We begin our work at gaining FimH and LRLO.
- FimH is from E.coli K12. We uses its genome as template and use PCR to gain FimH.
- As mentioned in "How to Attach in Intestine", there are five units in partJ36848 : a lacZ promoter, a ribosome binding site , Lpp aa 20, OmpA aa46-66 and streptavidin + His6 tag. Because we only need the first fout parts, so we use PCR to produce the sequence we need, LRLO or partC.(J36848 without streptavidin)
- The reverse primer of LRLO is designed complemented to the front primer of muFimH.
- Because there is a PstI cutting site in FimH, so we use a PCR method to mutate the cutting site. We desigh two set of primers, primerA(pA) anr primerB(pB).
- The front primer of pA and the reverse primer of pB is actually the primers we use to gain FimH, While pA reverse primer and pB front primer are designed to aneal to the two strand of PstI cutting site, while one of the base pair has been replaced.
- We use pA and pB to gained the front part of FimH as well as back part seperately. Because the reverse pA and front pB are complemented, so the two parts can be linked together by PCR, and since one of the basepair has been replaceed, so the cutting site will be mutated, thus we gain mutant FimH(muFimH, we also call it partD).
- After we gained muFimH and LRLO , we use PCR to link the two parts as the primer are complemented. Finally we get LRLO-muFimH sequence(partE), the main constructed sequence in our project.
- Before combining with other group's design, we need to test ours. Therefore we do two binding assay:
- One use GFP-partE to observe wether the ability of LRLO-FimH hybrids can attach to intestine.
- The other use pH sensor to replace lacZ promoter to see wether the protein expression could be regulate by pH.
Attachment Circuit Design
Detachment Circuit Design
Experiment Result
- Protocal and Method Check out
References
- Functional characterization of the FimH adhesin from Salmonella enterica serovar Enteritidis(D. Kisiela etc,2006)[http://mic.sgmjournals.org/cgi/content/full/152/5/1337?view=long&pmid=16622051]
- Combining sites of bacterial fimbriae(H. D. Grevea, L. Wynsa, J. Bouckaerta,2007) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6VS6-4PGPKS8-2&_user=1576506&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000053839&_version=1&_urlVersion=0&_userid=1576506&md5=33dfc6974f43b7ece79e0e7b90044ee7]
- Probing the receptor recognition site of the FimH adhesin by fimbriae- displayed FimH-FocH hybrids[http://mic.sgmjournals.org/cgi/reprint/144/7/1919?view=reprint&pmid=9695925]
- Host Protein Binding and Adhesive Properties of H6 and H7 Flagella of Attaching and Effacing Escherichia coli(A. L. Erdem etc,2007)[http://jb.asm.org/cgi/content/full/189/20/7426]
- Sequence Diversity of the Escherichia coli H7 fliC Genes:Implication for a DNA-Based Typing Scheme forE. coli O157:H7(L. WANG etc,2007)[http://jcm.asm.org/cgi/reprint/38/5/1786.pdf]
- Exploitation of host cells by enteropathogenic Escherichia coli(B. A. Vallance and B. B. Finlay,2000)[http://cmgm.stanford.edu/micro/MI209/EPEC.pdf]
- The Cell-binding Domain of Intimin from Enteropathogenic Escherichia coli Binds to b1 Integrins(G. Dougan etc,1996)[http://www.jbc.org/cgi/reprint/271/34/20359]
- Intimin from enteropathogenic Escherichia coli mediates remodelling of the eukaryotic cell surface(A.D.Phillips etc,2000)[http://mic.sgmjournals.org/cgi/content/full/146/6/1333]
- Intervening with Urinary Tract Infections Using Anti-Adhesives Based on the Crystal Structure of the FimH–Oligomannose-3 Complex(A. Wellens etc,2008)[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2323111]
 "
"