Team:BCCS-Bristol/Modeling
From 2008.igem.org
(Difference between revisions)
Tgorochowski (Talk | contribs) (→Models) |
Tgorochowski (Talk | contribs) (→Models) |
||
| Line 33: | Line 33: | ||
{| align="center" width="90%" | {| align="center" width="90%" | ||
| - | |width="33%" padding="3px"| <center>[[Image:BCCS-Modelling-GRN_Sec.jpg]]</center> | + | |width="33%" padding="3px"| <center>[[Team:BCCS-Bristol/Modeling-GRN|[[Image:BCCS-Modelling-GRN_Sec.jpg]]]]</center> |
|width="33%" padding="3px"| <center>[[Image:BCCS-Modelling-Agent_Sec.jpg]]</center> | |width="33%" padding="3px"| <center>[[Image:BCCS-Modelling-Agent_Sec.jpg]]</center> | ||
|width="33%" padding="3px"| <center>[[Image:BCCS-Modelling-Hybrid_Sec.jpg]]</center> | |width="33%" padding="3px"| <center>[[Image:BCCS-Modelling-Hybrid_Sec.jpg]]</center> | ||
Revision as of 21:09, 12 August 2008
|
Contents |
Approach
Modelling Notebook
- Progress Report - To track our progress see our weekly reports which let you know what we have been up to with comments on the reasons behind any important decisions.
- Modelling Parameters - Full listings of all parameters investigated for the modelling part of the project, with values that have been chosen, reasons for their selection and references to the original sources.
Models
 ]] ]] |  |  |
| | | |
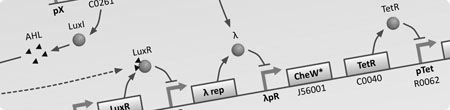
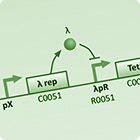
| To understand the dynamics of genetic networks that we design differental equations are used to model the changes in concentrations of mRNA and proteins. | With the project requiring global behaviour to emerge via the physical en masse movement of bacteria, stochastic agent based simulations are used to investigate the viability and limits of different approaches. | With the aim of improving simulation accuracy this model integrates the intercellular GRN dynamics with the extra-cellular interactions of the stochastic agent based simulation. |
Results
Modelling Photo Album
|
|
 "
"