Team:University of Lethbridge/Notebook/GeneralLabAugust
From 2008.igem.org
m (→Christa, Munima, Nathan Puhl, Roxanne) |
m (→August 16, 2008) |
||
| Line 92: | Line 92: | ||
| - | ===August | + | ===August 15, 2008=== |
====Roxanne==== | ====Roxanne==== | ||
| Line 98: | Line 98: | ||
Ran Plasmids in a 1% Agarose Gel at 100 V for 30 minutes. | Ran Plasmids in a 1% Agarose Gel at 100 V for 30 minutes. | ||
| + | |||
| + | ===August 16, 2008=== | ||
| + | |||
| + | ====Nathan Puhl, Roxanne==== | ||
| + | - | ||
| + | - | ||
===August 18=== | ===August 18=== | ||
Revision as of 21:37, 24 August 2008
(Aug. 1- 7 belong in other August notebooks)
Contents |
August 1, 2008
Nathan Puhl, Roxanne
Riboswitch 20 uL PCR. Set up 4 reactions (25 uL for each total volume). 1 uL or 1/100 pTopp and 1 uL or H1/100 PCR from July 28, 2008.
PCR conditions:
A. Initial denaturation: 98 C (3 min)
B. -Denaturation: 98 C (10 sec)
- Annealing: 55 C (30 sec)
-Extension: 72 C (15 sec)
-30 cycles
C. Final extension: 72 C (7 min)
August 2, 2008
Nathan Puhl, Roxanne
Ran riboswitch (Aug. 1, 2008) on 3% agarose gel. Results are in the hard copy lab notebook. Looks like 76 bp band will extract.
August 5, 2008
Nathan Puhl et al
Gel extracted two bands really closer to each other from PCR (August 1) at ~76 bp. Used Qiagen MiniElute kit. Results are in the hard copy lab notebook. Seems as if gel extraction was successful. ____ 2 color reporter system:
Set up digest of LacI and DT.
LacI - EcoRI and SpeI
DT - EcoRI and XbaI
Reaction mixture:
-5 uL template -5 uL NEB 2 Buffer -0.5 uL BSA? -1 uL RE #1 -1 uL RE #2 - 37.5 uL ddH20
Left at 37 C overnight. Heat deactivation (65 C) for 15 minutes.
August 6, 2008
Nathan Puhl, Roxanne
2 color reporter system:
Gel extracted LacI insert and DT vector. Didn't run gel yet.
August 7, 2008
Nathan Puhl, Roxanne
Riboswitch:
Set up PCR using purified riboswitch from Aug. 5, 2008 with platinum Taq (50 uL reaction). Made Master Mix for three reactions. Master Mix:
-10x Buffer (no Mg2+): 15 uL -10 mM dNTPs: 3 uL -50 mM Mg2+: 4.5 uL -10uM RF: 3 uL -10uM RR: 3 uL -Plat. poly: 0.6 iL -H20: 120.9 uL -template: 1 uL
Cycle conditions:
A. Initial denaturation: 94 C (2 min)
B. -Denaturation: 94 C (30 sec)
- Annealing: 55 C (30 sec)
-Extension: 72 C (30 sec)
-30 cycles
C. Final extension: 72 C (7 min)
Roxanne
-Ran the PCR Product on a 3% gel using only 1uL of DNA from the riboswitch.
August 13, 2008
Nathan Puhl and Sebastian
Verified and quantified the LacI and DT restriction digest products.
Set up a Ligase experiment for LacI and DT
- 1.3 uL vector(DT) - 8 uL insert(LacI) - 2 uL 10x Ligase buffer - 1.5 uL Ligase - 7.2 uL ddH2O - 20 uL total volume
August 14, 15
Nathan Puhl, Munima, Selina
Poured 61 LB + Amp plates. Stored in iGEM 4 C fridge.
August 15, 2008
Roxanne
Used Qiagen Plasmid MiniPrep Kit to Plasmid Prep Last Year's Biobrick Parts that were Incubated overnight.
Ran Plasmids in a 1% Agarose Gel at 100 V for 30 minutes.
August 16, 2008
Nathan Puhl, Roxanne
-
-
August 18
Christa, Nathan Puhl, Munima
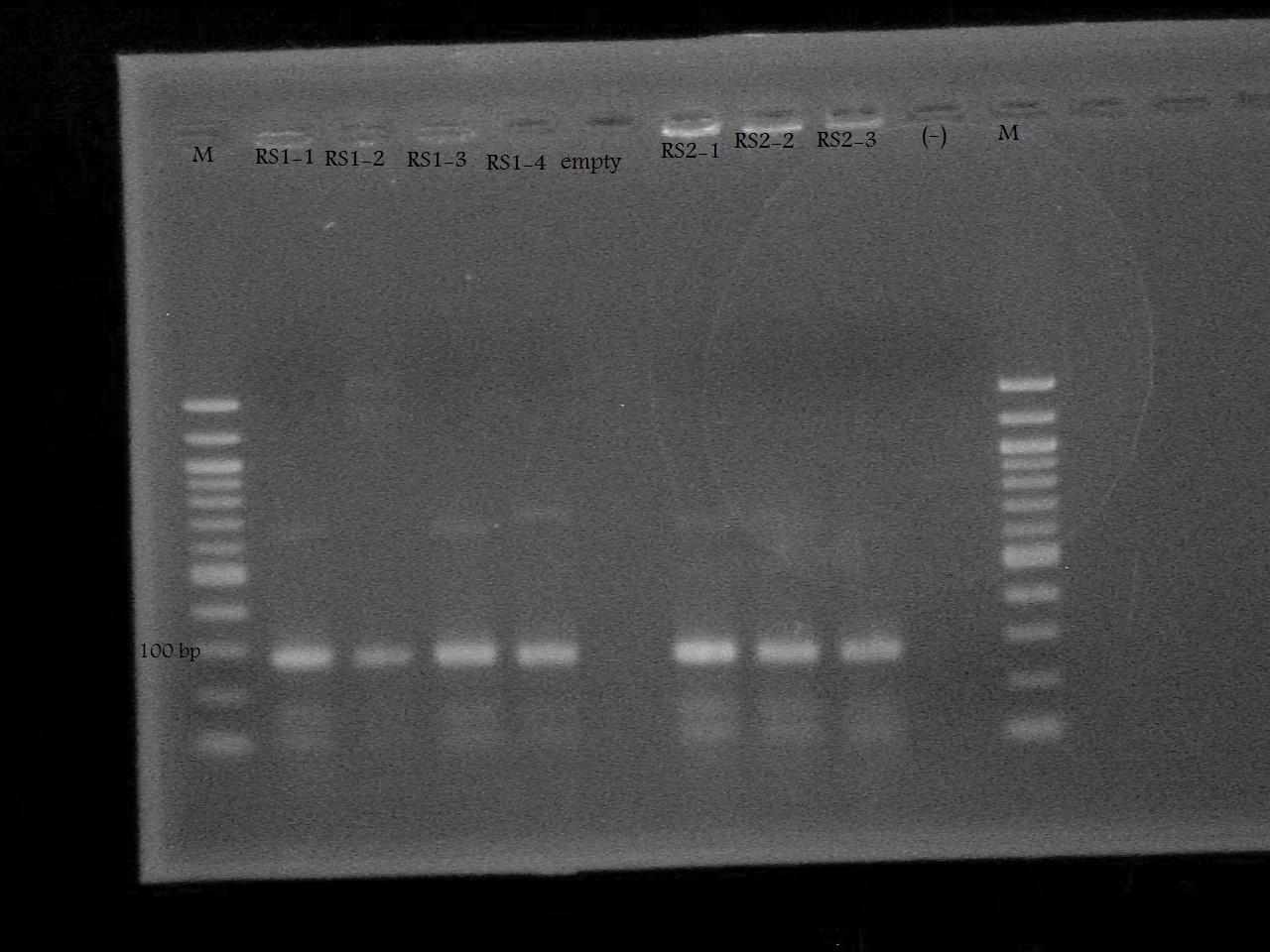
Ran a colony PCR of riboswitch extracted from gel done by Nathan and Roxanne on a previous day. Roxanne will remove the PCR tube from the thermocycler in the morning.
August 19, 2008
Roxanne
-Ran the PCR Product on a 2% Agarose Gel at 100 V for 33 minutes.
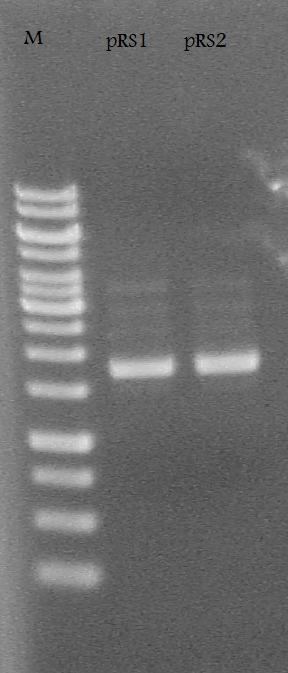
-Plasmid Prepped and made glycerol stocks from the RS1-1 and RS2-1 tubes of cells incubated in LB media + 100 ug/mL ampicillin.
Ran the pRS1 and pRS2 plasmids on a 1% Agarose Gel at 100 V for 30 minutes.
Christa, Munima, Sebastian
Did a plasmid prep on pSB1A7 using QIAprep Spin Miniprep Kit. Stocked 4- 50uL of pSB1A7 and stored in iGEM -20 C.
Sebastian, Nathan Puhl
Ran products from plasmid prep (pSB1A7 x 4 samples) on 1% agarose gel for 27 minutes.
-Lane 1 - 1 kb GeneRuler ladder (2 uL) -Lane 2 -6 pSB1A7 (3 uL) + 6x loading dye (2 uL); Mixed up what sample was in Lane 4, so Lanes 5 and 6 were run.
Conclusion: The bands appeared to be at the correct size for pSB1A7.
August 22, 2008
Christa, Munima, Nathan Puhl, Roxanne
Objective: Run a gel to confirm that appropriate inserts were amplified from the PCRs and do a gel extraction of the inserts to prepare them for the biobrick format.
-Could not obtain a picture of the gel (1% agarose) of the digested pSB1A7 and the recently amplified CheZ gene (15 uL x 3 wells) because the camera would not turn on. The CheZ gene appeared at the correct size (~700 bp). -Did a gel extraction of pSB1A7 and CheZ from that gel with the Qiagen MiniElute Gel Extraction kit. Final volume of each was 10 uL. -Ran another 1% agarose gel to confirm that the gel extraction was successful. Ran 1 uL of each sample. Bands appeared at appropriate sizes. The concentrations of pSB1A7 and CheZ were estimated to be 25 ng/uL and 80 ng/uL, respectively.
Next step: Digest both pSB1A7 and CheZ with phosphatase. A restriction digest will be performed at a later date on CheZ to prepare it for insertion.
 "
"