Team:Illinois/Antibody Receptor Tyrosine Kinase Fusion Notebook
From 2008.igem.org
| Line 63: | Line 63: | ||
6. Resuspend in 10 mL cold 30 mM CaCl2, incubated on ice overnight | 6. Resuspend in 10 mL cold 30 mM CaCl2, incubated on ice overnight | ||
| + | |||
| + | |||
| + | == July 19, 2008 == | ||
| + | Glycerol added to 10%, cells in freezer | ||
| + | |||
| + | Transformation: | ||
| + | |||
| + | 1. Cells put in ice 30 min with 1μL plasmid(100ng) | ||
| + | |||
| + | 2. Heat Shock for 2 minutes at 42 degrees | ||
| + | |||
| + | 3. Ice for 8 minutes | ||
| + | |||
| + | 4. Add cells to 1μL LB | ||
| + | |||
| + | 5. Grow for 1 hour | ||
| + | |||
| + | 6. Plate 100 to 200 μL on LB and amp, grow overnight | ||
| + | |||
| + | |||
| + | == July 21, 2008 == | ||
| + | Observations: | ||
| + | |||
| + | Plenty of colonies on all plates | ||
| + | |||
| + | Also, making 5mL overnight cultures for mini-prep tomorrow. | ||
| + | |||
| + | |||
| + | == July 22, 2008 == | ||
| + | Mini spin prep on LC and HC colonies | ||
| + | |||
| + | Place DNA in 100μL H2O in freezer | ||
| + | |||
| + | |||
| + | == July 29, 2008 == | ||
| + | All primers brought to standard concentration, 30mM | ||
| + | |||
| + | 33.3μL H2O added per n mole of primer | ||
| + | |||
| + | |||
| + | == July 30, 2008 == | ||
| + | Assembly PCR: amplify antibody gene fragments and then assemble them, light and heavy chains | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | |tube | ||
| + | |1 | ||
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |6 | ||
| + | |7 | ||
| + | |8 | ||
| + | |- | ||
| + | |Antibody chain | ||
| + | |H | ||
| + | |H | ||
| + | |L | ||
| + | |L | ||
| + | |H | ||
| + | |H | ||
| + | |L | ||
| + | |L | ||
| + | |- | ||
| + | |DNA source | ||
| + | |colspan="4"|mini-prep | ||
| + | |colspan="4"|synthesized IDT genes | ||
| + | |- | ||
| + | |template | ||
| + | |colspan="8"|0.5ul | ||
| + | |- | ||
| + | |primers | ||
| + | |colspan="8"|1ul of appropriate forward and reverse primer | ||
| + | |- | ||
| + | |MgCl2 | ||
| + | |3ul | ||
| + | |5ul | ||
| + | |3ul | ||
| + | |5ul | ||
| + | |3ul | ||
| + | |5ul | ||
| + | |3ul | ||
| + | |5ul | ||
| + | |- | ||
| + | |master mix | ||
| + | |colspan="8"|20ul | ||
| + | |- | ||
| + | |H20 | ||
| + | |colspan="8"|to 50ul total volume | ||
| + | |} | ||
| + | |||
| + | Master mix already contains MgCl2; should have added extra to some tubes. | ||
| + | |||
| + | PCR program: 1. 94 degrees 5 min | ||
| + | 2. 94 degrees 1 min | ||
| + | 3. 50 degrees 1 min | ||
| + | 4. 72 degrees 1 min | ||
| + | 5. GOTO 2. 39 cycles | ||
| + | 6. HOLD at 4 degrees | ||
| + | |||
| + | 1.5% agarose gel made, 0.75g agarose in 50ml 0.5X TBE w/10ul EtBr. | ||
| + | |||
| + | 20ul of PCR produts loaded on gel w/4ul 6X loading buffer, run at 200V for about half an hour. | ||
| + | |||
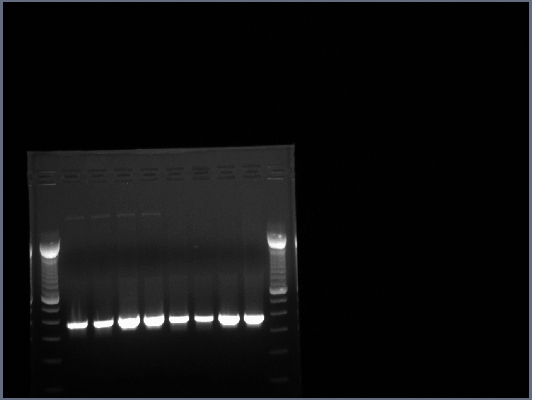
| + | All lanes had lots of DNA at ~375bp, PCR worked. | ||
| + | [[Image:07-30-08_AbFrag_Gel1.jpg|none]] | ||
<!-- == Insert Date Here == | <!-- == Insert Date Here == | ||
* lab procedure | * lab procedure | ||
Revision as of 01:33, 8 October 2008
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
Contents |
July 1, 2008
Made Yeast Media
YPD Medium, per liter:
10g yeast extract
20g peptone
20g dextrose
20g agar(only for plates)
1. Dextrose filter sterilized, the rest autoclaved
2. Weigh nutrients into flask double the volume you want to make, and stir to dissolve
3. Dextrose added to autoclaved media to equivalent of 20 g/L
4. Liquid media placed on bench, plate media placed in 65 degree water bath approximately 5 minutes
5. Poured into plates and allowed to solidify
July 17, 2008
E.Coli Media
LB medium, per liter
10g tryptone
5g yeast extract
5g NaCl
1 mL 1N NaOH
15g (agar for plates)
1. Antibody DNA resuspended in TE buffer, 0.1 μg/μL
2. 5mL LB inoculated with single colony DH5a pro
3. Incubated at 37 degrees overnight
July 18, 2008
Competent cells:
1. 3mL overnight culture of DH5a pro
2. Inoculated into 35mL of LB
3. OD600 checked, want 0.2-0.3
4. Place culture on ice for 3 minutes
5. Spin at 10,000 rpm for 7 minutes, discard supernatant
6. Resuspend in 10 mL cold 30 mM CaCl2, incubated on ice overnight
July 19, 2008
Glycerol added to 10%, cells in freezer
Transformation:
1. Cells put in ice 30 min with 1μL plasmid(100ng)
2. Heat Shock for 2 minutes at 42 degrees
3. Ice for 8 minutes
4. Add cells to 1μL LB
5. Grow for 1 hour
6. Plate 100 to 200 μL on LB and amp, grow overnight
July 21, 2008
Observations:
Plenty of colonies on all plates
Also, making 5mL overnight cultures for mini-prep tomorrow.
July 22, 2008
Mini spin prep on LC and HC colonies
Place DNA in 100μL H2O in freezer
July 29, 2008
All primers brought to standard concentration, 30mM
33.3μL H2O added per n mole of primer
July 30, 2008
Assembly PCR: amplify antibody gene fragments and then assemble them, light and heavy chains
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Antibody chain | H | H | L | L | H | H | L | L |
| DNA source | mini-prep | synthesized IDT genes | ||||||
| template | 0.5ul | |||||||
| primers | 1ul of appropriate forward and reverse primer | |||||||
| MgCl2 | 3ul | 5ul | 3ul | 5ul | 3ul | 5ul | 3ul | 5ul |
| master mix | 20ul | |||||||
| H20 | to 50ul total volume | |||||||
Master mix already contains MgCl2; should have added extra to some tubes.
PCR program: 1. 94 degrees 5 min 2. 94 degrees 1 min 3. 50 degrees 1 min 4. 72 degrees 1 min 5. GOTO 2. 39 cycles 6. HOLD at 4 degrees
1.5% agarose gel made, 0.75g agarose in 50ml 0.5X TBE w/10ul EtBr.
20ul of PCR produts loaded on gel w/4ul 6X loading buffer, run at 200V for about half an hour.
All lanes had lots of DNA at ~375bp, PCR worked.
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
 "
"