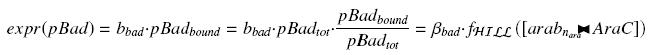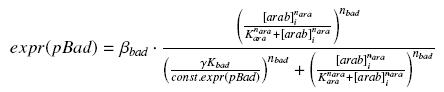Team:Paris/Modeling/f2
From 2008.igem.org
(Difference between revisions)
| Line 26: | Line 26: | ||
|min<sup>-1</sup> | |min<sup>-1</sup> | ||
|0.0198 | |0.0198 | ||
| + | | | ||
|- | |- | ||
|[GFP] | |[GFP] | ||
Revision as of 11:40, 12 October 2008
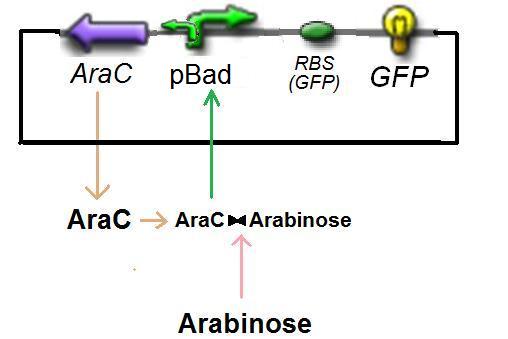
The experience would give us
Thus, at steady-state and in the exponential phase of growth :
| param | signification | unit | value | comments |
| [expr(pBad)] | expression rate of pBad with RBS E0032 | nM.min-1 | need for 20 measures with well choosen [arab]i | |
| γGFP | dilution-degradation rate of GFP(mut3b) | min-1 | 0.0198 | |
| [GFP] | GFP concentration at steady-state | nM | need for 20 measures | |
| (fluorescence) | value of the observed fluorescence | au | need for 20 measures | |
| conversion | conversion ratio between fluorescence and concentration | nM.au-1 | (1/79.429) |
| param | signification corresponding parameters in the equations | unit | value | comments |
| βbad | production rate of pBad with RBS E0032 not in the system | nM.min-1 | ||
| (γ Kbad/const.expr(pBad)) | activation constant of pBad not in the system | nM | ||
| nbad | complexation order of pBad not in the system | no dimension | The literature [?] gives nbad = | |
| Kara | complexation constant Arabinose-AraC not in the system | nM | The literature [?] gives Kara = | |
| nara | complexation order Arabinose-AraC not in the system | no dimension | The literature [?] gives nara = |
That will give us directly ƒ2([arab])
 "
"