Team:Imperial College/Genetic Circuit
From 2008.igem.org
m |
m |
||
| Line 35: | Line 35: | ||
}} | }} | ||
| - | {{Imperial/Box1|Modelling Inducible Gene Expression | + | {{Imperial/Box1|Modelling Inducible Gene Expression| |
| - | |[[Image:Phase 2-linduced.PNG|thumb|300px]] | + | [[Image:Phase 2-linduced.PNG|thumb|300px]] |
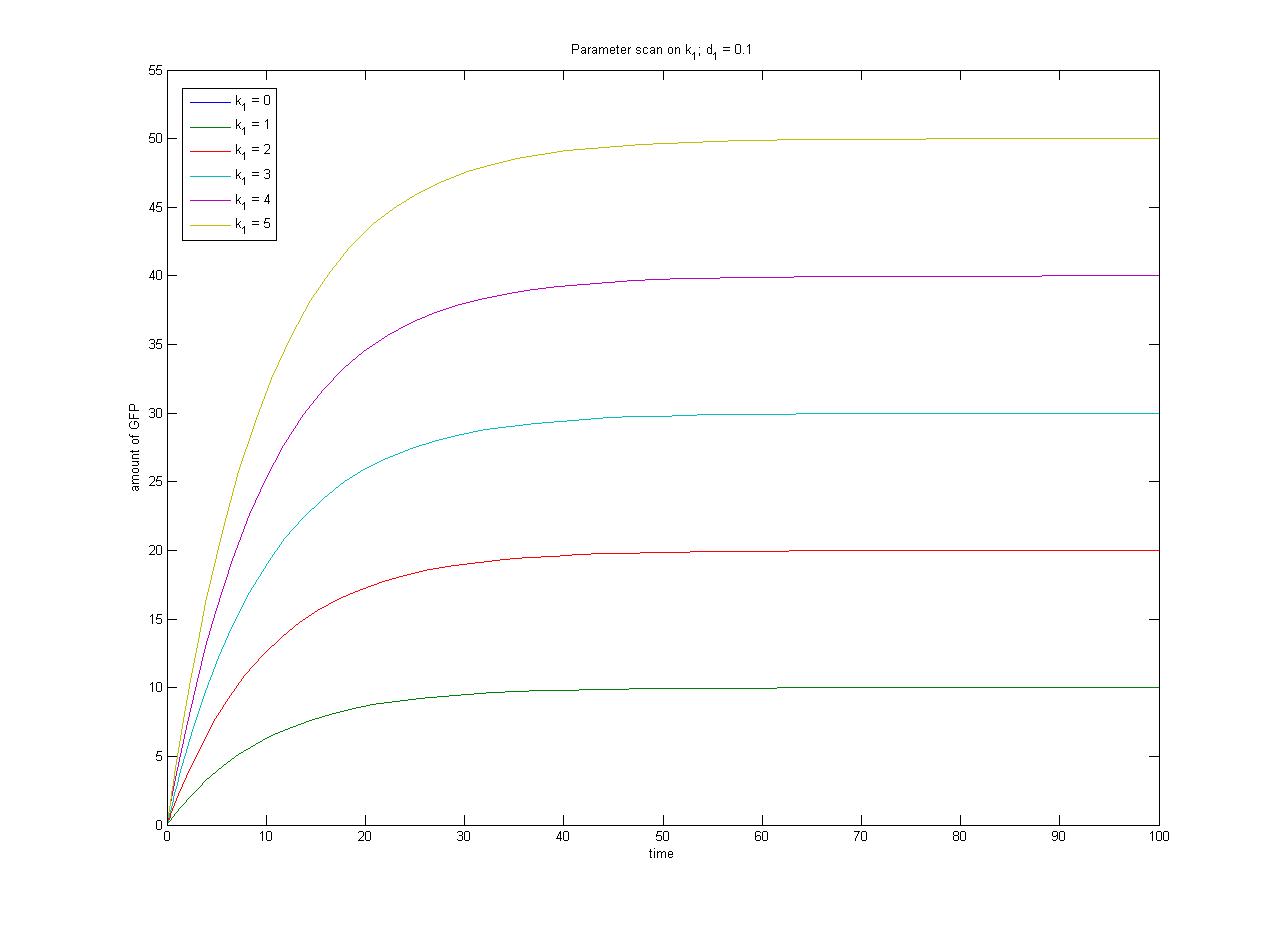
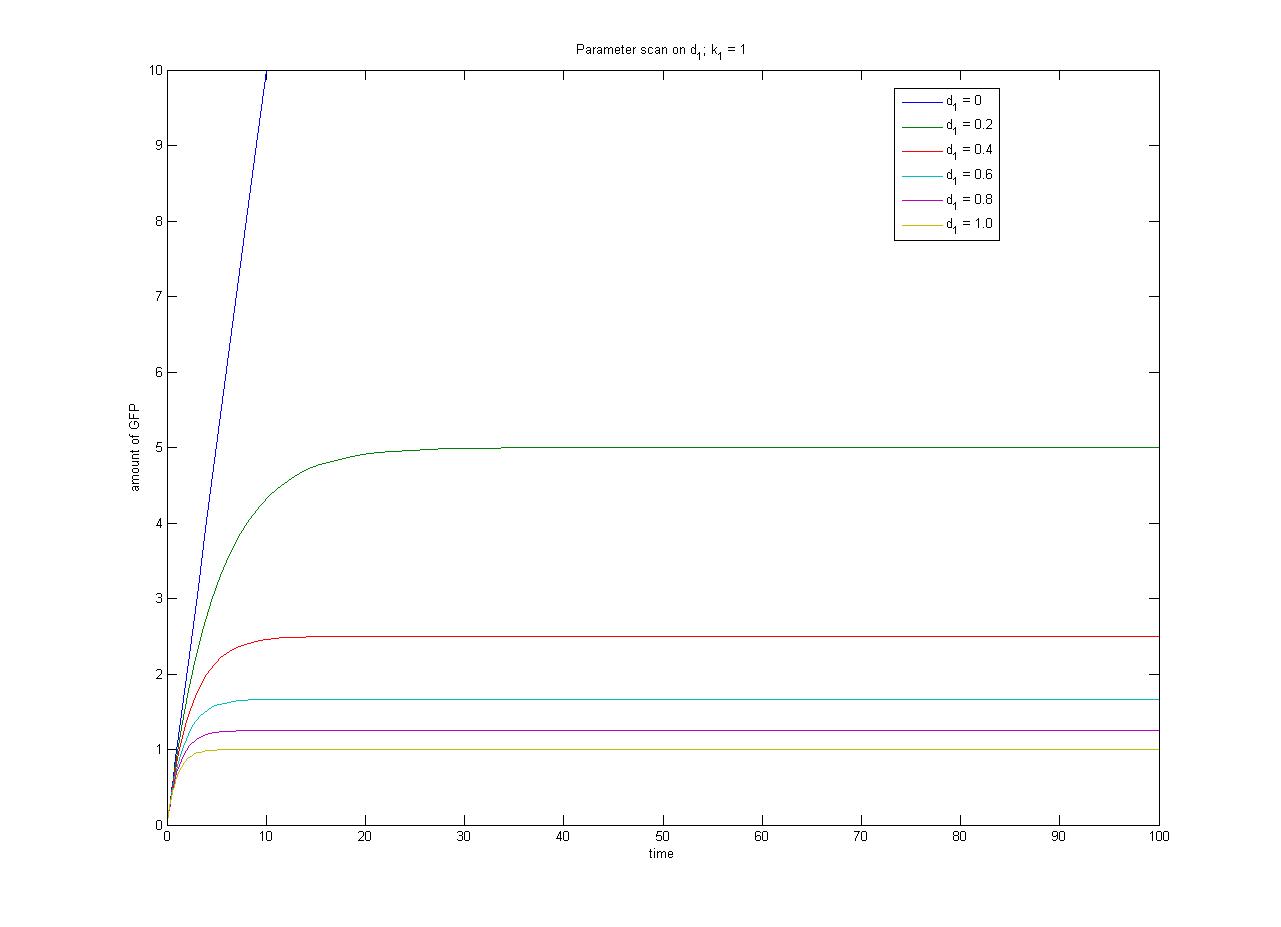
The repressor is constitutively expressed. Hence we can assume the constitutive expression model from the previous characterisation step. | The repressor is constitutively expressed. Hence we can assume the constitutive expression model from the previous characterisation step. | ||
Revision as of 17:01, 13 October 2008
{{Imperial/Box1|Modelling Inducible Gene Expression| The repressor is constitutively expressed. Hence we can assume the constitutive expression model from the previous characterisation step. <math>\frac{d[R]}{dt} = k_{1} - d_{1}[R]</math> When the inducer is added it binds reversibly to the repressor. <math> R + I \Leftrightarrow RI </math> Repressor only binds to the promoter when it is in its unbound form, hence transcription will be a function of free repressor concentration. <math> Transcription = \frac{\beta.{[R]}^n}{{K_m}^n+[R]^n}</math> And overall protein expression can be described as <math>\frac{d[protein]}{dt} = Transcription - d_2[protein]</math> The ODEs and simulation may be found in the Appendices section of the Dry Lab hub. <biblio>
</biblio> }} |
|||||||||||||
 "
"