Team:ETH Zurich/Modeling/Overview
From 2008.igem.org
Luca.Gerosa (Talk | contribs) |
Luca.Gerosa (Talk | contribs) |
||
| Line 48: | Line 48: | ||
* Which are the predicted quantitative differences in terms of growth rate and genome size of strains on which has been applied the selection procedure? | * Which are the predicted quantitative differences in terms of growth rate and genome size of strains on which has been applied the selection procedure? | ||
<br><br> | <br><br> | ||
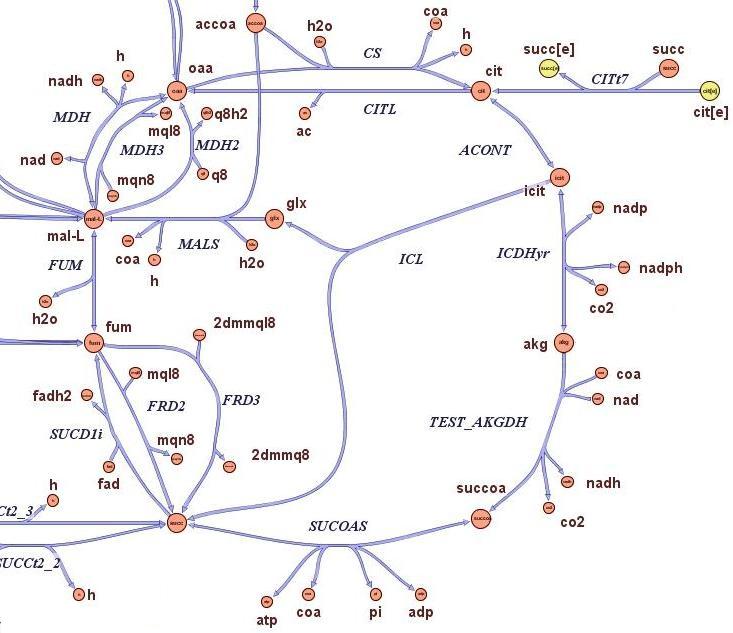
| - | '''Method:''' <br> | + | '''Method:''' The state-of-the-art genome scale model for E.Coli MG1655 has been modified in order to account for thymidine auxotrophycity, thymidine uptaking limitation, genome reduction and growth in different medium. Flux balance analysis has been used to determine growth rates.<br> |
'''Results:''' <br> | '''Results:''' <br> | ||
</div> | </div> | ||
Revision as of 16:03, 11 October 2008
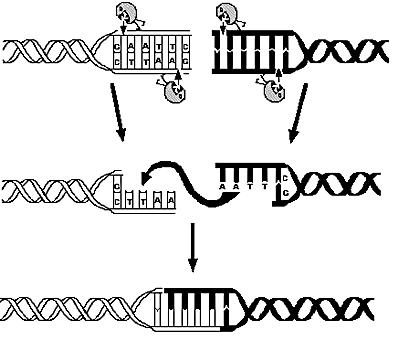
Overview on the modelling frameworkThis page is meant to give an introduction to the the overall modelling framework we have constructed in order to asses feasibility analysis, temporal scale details and other parameter estimations that regard our project setup. As introduced in the project overview section, four main components can be identified in the deviced mechanism. Accordingly, we divided the modelling framework in four modules that tackles the relative problematics.
|
 "
"