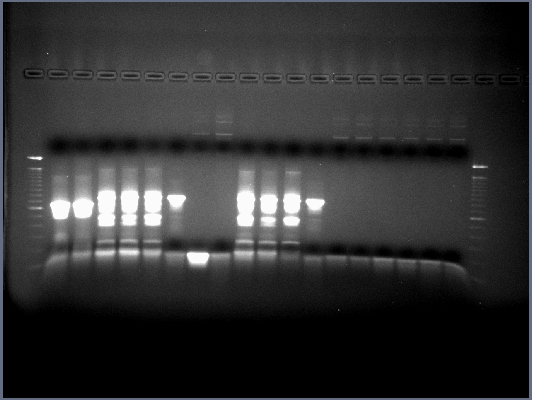
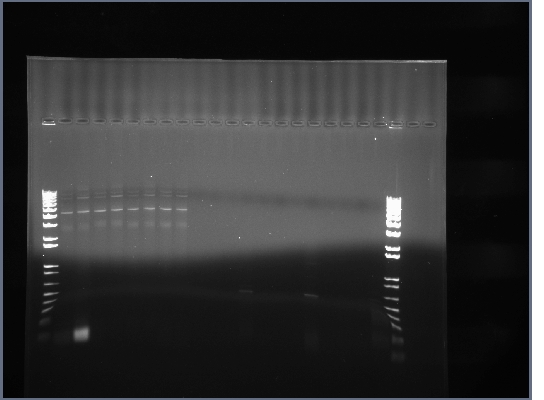
Team:Illinois/Antibody Receptor Tyrosine Kinase Fusion Notebook
From 2008.igem.org
| Line 1: | Line 1: | ||
{{bottom_template}} | {{bottom_template}} | ||
| - | == | + | == July 1, 2008 == |
Made Yeast Media | Made Yeast Media | ||
| Line 25: | Line 25: | ||
| - | == | + | == July 17, 2008 == |
E.Coli Media | E.Coli Media | ||
| Line 47: | Line 47: | ||
| - | == | + | == July 18, 2008 == |
Competent cells: | Competent cells: | ||
| Line 64: | Line 64: | ||
7. Glycerol added to 10%, cells in freezer | 7. Glycerol added to 10%, cells in freezer | ||
| - | == | + | |
| + | == July 19, 2008 == | ||
Transformation: | Transformation: | ||
| Line 81: | Line 82: | ||
| - | == | + | == July 21, 2008 == |
Observations: | Observations: | ||
| Line 89: | Line 90: | ||
| - | == | + | == July 22, 2008 == |
Mini spin prep on LC and HC colonies | Mini spin prep on LC and HC colonies | ||
| Line 95: | Line 96: | ||
| - | == | + | == July 29, 2008 == |
All primers brought to standard concentration, 30mM | All primers brought to standard concentration, 30mM | ||
| Line 101: | Line 102: | ||
| - | == | + | == July 30, 2008 == |
Assembly PCR: amplify antibody gene fragments and then assemble them, light and heavy chains | Assembly PCR: amplify antibody gene fragments and then assemble them, light and heavy chains | ||
| Line 171: | Line 172: | ||
| - | == | + | == July 31, 2008 == |
PCR to asemble fragments from yesterday. | PCR to asemble fragments from yesterday. | ||
| Line 221: | Line 222: | ||
| - | == | + | == August 1, 2008 == |
PCR troubleshooting of yesterday's reaction: | PCR troubleshooting of yesterday's reaction: | ||
use gradient for annealing temp (40-65 degrees), use more template, leave out end primers | use gradient for annealing temp (40-65 degrees), use more template, leave out end primers | ||
| Line 253: | Line 254: | ||
|} | |} | ||
| - | + | Annealing temps: 40, 43.8, 50, 54, 60, and 64 degrees | |
PCR program | PCR program | ||
| Line 270: | Line 271: | ||
| - | == | + | == August 6, 2008 == |
1.5% agarose gel of 8-1 PCR products run to cut out bands. Products A1, B1, C1, and E1 used. | 1.5% agarose gel of 8-1 PCR products run to cut out bands. Products A1, B1, C1, and E1 used. | ||
| Line 276: | Line 277: | ||
| - | == | + | == September 5, 2008 == |
Flk-1 clones from [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=MGC-18600&Template=mgcMouseClones ATCC] streaked onto LB+amp and incubated at 37 degrees. | Flk-1 clones from [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=MGC-18600&Template=mgcMouseClones ATCC] streaked onto LB+amp and incubated at 37 degrees. | ||
| - | == | + | == September 6, 2008 == |
Colonies picked from flk-1 plate and inoculated into 5ml LB+amp, grown at 37 degrees. | Colonies picked from flk-1 plate and inoculated into 5ml LB+amp, grown at 37 degrees. | ||
| Line 286: | Line 287: | ||
| - | == | + | == September 7, 2008 == |
Mini spin prep of flk-1 and YCp50-poly cells, plasmids stored in 50ul H2O. | Mini spin prep of flk-1 and YCp50-poly cells, plasmids stored in 50ul H2O. | ||
| - | == | + | == September 25, 2008 == |
Biobricks | Biobricks | ||
| Line 412: | Line 413: | ||
# HOLD at 4 degrees | # HOLD at 4 degrees | ||
| - | == | + | == September 26, 2008 == |
*product from yesterday run on 1% gel, 200V for ~30 minutes | *product from yesterday run on 1% gel, 200V for ~30 minutes | ||
| Line 488: | Line 489: | ||
# HOLD at 4 degrees | # HOLD at 4 degrees | ||
| - | == | + | == September 27, 2008 == |
*0.8% agarose gel run of products from yesterday, 200V for ~45 minutes | *0.8% agarose gel run of products from yesterday, 200V for ~45 minutes | ||
| Line 630: | Line 631: | ||
== October 17, 2008 == | == October 17, 2008 == | ||
| - | Transformation failed | + | Transformation failed. |
1% gel of yesterday's PCR run, 200V, ~40min, no bands | 1% gel of yesterday's PCR run, 200V, ~40min, no bands | ||
Revision as of 20:27, 18 October 2008
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
July 1, 2008
Made Yeast Media
YPD Medium, per liter:
10g yeast extract
20g peptone
20g dextrose
20g agar(only for plates)
1. Dextrose filter sterilized, the rest autoclaved
2. Weigh nutrients into flask double the volume you want to make, and stir to dissolve
3. Dextrose added to autoclaved media to equivalent of 20 g/L
4. Liquid media placed on bench, plate media placed in 65 degree water bath approximately 5 minutes
5. Poured into plates and allowed to solidify
July 17, 2008
E.Coli Media
LB medium, per liter
10g tryptone
5g yeast extract
5g NaCl
1 mL 1N NaOH
15g (agar for plates)
1. Antibody DNA resuspended in TE buffer, 0.1 μg/μL
2. 5mL LB inoculated with single colony DH5a pro
3. Incubated at 37 degrees overnight
July 18, 2008
Competent cells:
1. 3mL overnight culture of DH5a pro
2. Inoculated into 35mL of LB
3. OD600 checked, want 0.2-0.3
4. Place culture on ice for 3 minutes
5. Spin at 10,000 rpm for 7 minutes, discard supernatant
6. Resuspend in 10 mL cold 30 mM CaCl2, incubated on ice overnight
7. Glycerol added to 10%, cells in freezer
July 19, 2008
Transformation:
1. Cells put in ice 30 min with 1μL plasmid(100ng)
2. Heat Shock for 2 minutes at 42 degrees
3. Ice for 8 minutes
4. Add cells to 1ml LB
5. Grow for 1 hour
6. Plate 100 to 200 μL on LB and amp, grow overnight
July 21, 2008
Observations:
Plenty of colonies on all plates
Also, making 5mL overnight cultures for mini-prep tomorrow.
July 22, 2008
Mini spin prep on LC and HC colonies
Place DNA in 100μL H2O in freezer
July 29, 2008
All primers brought to standard concentration, 30mM
33.3μL H2O added per n mole of primer
July 30, 2008
Assembly PCR: amplify antibody gene fragments and then assemble them, light and heavy chains
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Antibody chain | H | H | L | L | H | H | L | L |
| DNA source | mini-prep | synthesized IDT genes | ||||||
| template | 0.5ul | |||||||
| primers | 1ul of appropriate forward and reverse primer | |||||||
| MgCl2 | 3ul | 5ul | 3ul | 5ul | 3ul | 5ul | 3ul | 5ul |
| master mix | 20ul | |||||||
| H20 | to 50ul total volume | |||||||
Master mix already contains MgCl2; should have added extra to some tubes.
PCR program
- 94 degrees 5 min
- 94 degrees 1 min
- 50 degrees 1 min
- 72 degrees 1 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
1.5% agarose gel made, 0.75g agarose in 50ml 0.5X TBE w/10ul EtBr.
20ul of PCR produts loaded on gel w/4ul 6X loading buffer, run at 200V for about half an hour.
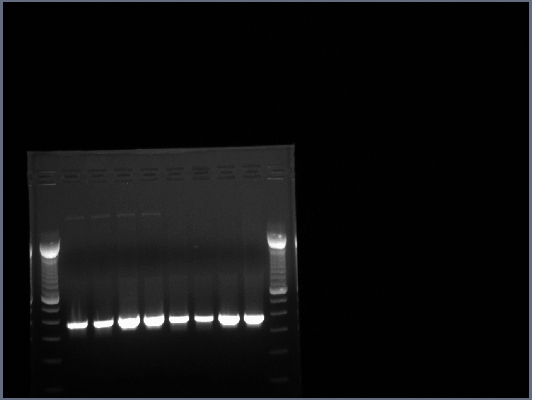
All lanes had lots of DNA at ~375bp, PCR worked.
July 31, 2008
PCR to asemble fragments from yesterday.
| tube | 1 | 2 | 3 | 4 | ||||
| template from 7-30 PCR | 2+3 | 2+7 | 6+7 | 1+8 | ||||
| template | 0.5ul of both heavy and light chains | |||||||
| primers | 1ul of appropriate forward and reverse primer | |||||||
| MgCl2 | 0ul | 0ul | 3ul | 3ul | ||||
| master mix | 20ul | |||||||
| H20 | to 50ul total volume | |||||||
PCR program
- 94 degrees 5 min
- 94 degrees 1 min
- 50 degrees 1 min
- 72 degrees 1 min
- GOTO 2. 29 cycles
- 72 degrees 5 min
- HOLD at 4 degrees
Products run on a 1.5% agarose gel, no products of 700-800bp.
August 1, 2008
PCR troubleshooting of yesterday's reaction: use gradient for annealing temp (40-65 degrees), use more template, leave out end primers
Four series of reactions using a gradient annealing temp were run in addition to a fifth series omitting the end primers.
| reaction series | A | B | C | D | E |
| template (5+7) from 7-30 PCR | 0.5ul | 1ul | 1.5ul | 2ul | 1ul |
| primers | 1ul of forward and reverse primers (omitted in E series) | ||||
| master mix | 20ul | ||||
| H20 | to 50ul total volume | ||||
Annealing temps: 40, 43.8, 50, 54, 60, and 64 degrees
PCR program
- 94 degrees 5 min
- 94 degrees 1 min
- annealing step 1 min
- 72 degrees 1 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
PCR products run on 1.5% agarose gel at 200V for ~20 minutes, then 240V for ~20 minutes.
Products in the 700-800bp range in all lanes. Perhaps it worked because of using a differnt product from the 7-30 PCR.
August 6, 2008
1.5% agarose gel of 8-1 PCR products run to cut out bands. Products A1, B1, C1, and E1 used.
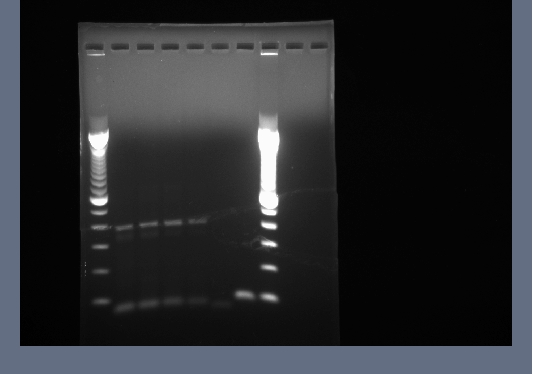
Bands around 800bp cut out for all lanes and invitrogen gel extraction kit used to collect DNA in 50ul H2O.
September 5, 2008
Flk-1 clones from [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=MGC-18600&Template=mgcMouseClones ATCC] streaked onto LB+amp and incubated at 37 degrees.
September 6, 2008
Colonies picked from flk-1 plate and inoculated into 5ml LB+amp, grown at 37 degrees.
Freeze-dried e. coli with YCp50-poly from [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=87555&Template=vectors ATCC] resuspended in 5ml LB, grown at 37 degrees.
September 7, 2008
Mini spin prep of flk-1 and YCp50-poly cells, plasmids stored in 50ul H2O.
September 25, 2008
Biobricks
Bricks to be made: H chain, L chain, SCA, flk-1
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| 0.5uL template | H chain | L chain | SCA | SCA | SCA | SCA | flk-1 | flk-1 |
| IDT | IDT | A | B | C | E | A | B | |
| primers | 1uL forward & reverse for all | |||||||
| mastermix | 8.25uL for all | |||||||
| H2O | 39.25uL for all | |||||||
| tube | 9 | 10 | 11 | 12 | 13 | 14 | ||
| 0.5uL template | SCA w/ link | SCA w/ link | SCA w/ link | SCA w/ link | flk link 1 | flk link 1 | ||
| A | B | C | E | A | B | |||
| primers | 1uL forward & reverse for all | |||||||
| mastermix | 8.25uL for all | |||||||
| H2O | 39.25uL for all | |||||||
| tube | 15 | 16 | 17 | 18 | ||||
| 0.5uL template | flk link 2 | flk link 2 | flk link 3 | flk link 3 | ||||
| A | B | A | B | |||||
| primers | 1uL forward & reverse for all | |||||||
| mastermix | 8.25uL for all | |||||||
| H2O | 39.25uL for all | |||||||
PCR program
- 94 degrees 4 min
- 94 degrees 1 min
- 50 degrees 1 min
- 72 degrees 3 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
September 26, 2008
- product from yesterday run on 1% gel, 200V for ~30 minutes
- mostly good results, small amound of DNA for flk-1
- lanes 1,2,6,8,12,14,16,18 cut out and purified with invitrogen gel extraction kit, eluted in 50ul H2O
PCR to assemble SCA-flk-1 fusion and increase flk-1 yield
| tube | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| template | 0.5ul flk-1 A | 0.5ul flk-1 B | 0.5ul flk link 1 A | 0.5ul flk link 1 B | 0.5ul flk link 2 A | 0.5ul flk link 2 B | 0.5ul flk link 3 A | 0.5ul flk link 3 B | 0.5ul SCA link + 0.5ul flk link 1 | 0.5ul SCA link + 0.5ul flk link 1 (crude) | 0.5ul SCA link + 2ul flk link 1 | 0.5ul SCA link + 2ul flk link 1 (crude) | 0.5ul SCA link + 0.5ul flk link 2 | 0.5ul SCA link + 0.5ul flk link 2 (crude) | 0.5ul SCA link + 2ul flk link 2 | 0.5ul SCA link + 2ul flk link 2 (crude | 0.5ul SCA link + 0.5ul flk link 3 | 0.5ul SCA link + 0.5ul flk link 3 (crude) | 0.5ul SCA link + 2ul flk link 3 | 0.5ul SCA link + 2ul flk link 3 (crude) |
| primers | 1uL forward & reverse for all | |||||||||||||||||||
| mastermix | 8.25uL for all | |||||||||||||||||||
| H2O | to 50ul total volume | |||||||||||||||||||
PCR program
- 94 degrees 4 min
- 94 degrees 1 min
- 50 degrees 1 min
- 72 degrees 3 min
- GOTO 2. 29 cycles
- HOLD at 4 degrees
September 27, 2008
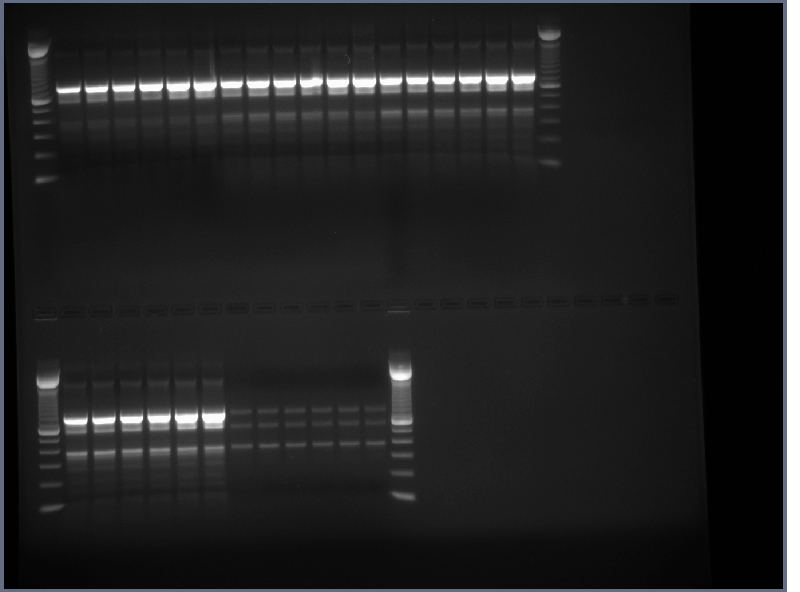
- 0.8% agarose gel run of products from yesterday, 200V for ~45 minutes
- first 8 reactions may have worked but bands not great, assembly reactions did not work
September 28, 2008
0.8% gel run of yesterday's products, 150 V, 38 min, 200 V, 10 min
September 30, 2008
p1010 plasmid for biobicks cut out of plate 1003, well 4A; soacked in 5uL warm TE (50°C) for ~20 min, heat shocked at 50°C for 60s, placed on ice 2 min, 200uL LB added, incubated at 37°C, 1 hour, plated on LB + Amp and incubated overnight.
Restriction digest of our biobricks 18uL H2O 2uL 10x buffer Z 1uL EcoRI and PstI 1,2,4 uL DNA (H chain, L chain, SCA, flk)
p1010 transformatoin was stupid, transformed another plasmid instead, 1008, 1C
| tube | 1 | 2 | 3 | 4 | 5 | 6 | ||
| template | A=0.5uL B=1.0uL C=1.5uL | (all flk-1 B) | ||||||
| primers | 1uL forward & reverse for all | |||||||
| mastermix | 8.25uL for all | |||||||
| H2O | 39.25uL for A | 38.75uL for B | 38.25uL for C | |||||
| tube | 1 | 2 | 3 | 4 | 5 | 6 | ||
| gradient | 55.0 | 57.8 | 59.3 | 61.0 | 63.5 | 65.0 | ||
| temps | (1) | (5) | (6) | (7) | (9) | (12) | ||
Run for 39 cycles
October 2, 2008
PCR products run on 0.8% gel, 200V, ~40min Same results as always
pSB1A3 & pSB1A7 cut out of registry and transformed Plated on LB+Amp
October 3, 2008
PCR from yesterday may have kind of worked, just not a lot of product, running another gel to find out
Transformation failed, transforming w/ RDR plasmid to test transformation protocol
Hard to tell if PCR products are correct length, so-so bands
October 8, 2008
Transformations to get plasmids 1003:2F, 1008:10D, 1008:1C, 1008:6G
October 10, 2008
DNA purification using S prime PerfectPrep(TM) Spin Mini kit on transformed bacteria from 10/8/2008
October 14, 2008
Transformation using plasmid from 10/10/2008
October 16, 2008
Transformation failed, duh top 10 50uL cells transformed with 1003:10D, 1003:2F, plasmid from 10/10/2008, and pUC19 control
PCR to make ARTK fusion
| tube | 1 | 2 | 3 | 4 | 5 |
| template | 0.5uL SCA + 0.5uL flk-link3(8) | 0.5uL SCA + 1.0uL flk-link3(8) | 0.5uL SCA + 1.5uL flk-link3(8) | 0.5uL SCA + 2.0uL flk-link3(8) | 0.5uL SCA + 1.5uL flk-link3(8) |
| primers | 1.0uL forward & reverse | none | |||
| mastermix | 20.0uL for all | ||||
| H2O | 27.0uL | 26.5uL | 26.0uL | 25.5uL | 28.0uL |
October 17, 2008
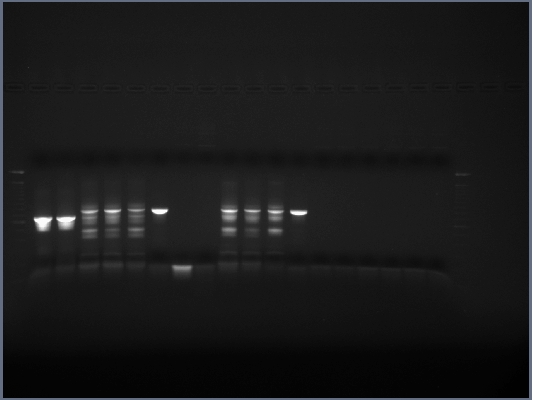
Transformation failed. 1% gel of yesterday's PCR run, 200V, ~40min, no bands
October 18, 2008
Using phusion to get ARTK
| tube | 1 | 2 | 3 | 4 | 5 |
| template | 0.5uL SCA + 0.5uL flk-link3(8) | 0.5uL SCA + 1.0uL flk-link3(8) | 0.5uL SCA + 1.5uL flk-link3(8) | 0.5uL SCA + 2.0uL flk-link3(8) | 0.5uL SCA + 1.5uL flk-link3(8) |
| primers | 1.0uL forward & reverse | no primers | |||
| buffer | 10.0uL for all | ||||
| phu | 0.5uL for all | ||||
| H2O | 36.5uL | 36.0uL | 35.5uL | 35.0uL | 37.5uL |
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
 "
"