Team:Imperial College/Genetic Circuit
From 2008.igem.org
m |
m |
||
| Line 5: | Line 5: | ||
{{Imperial/Box1|Modelling Constitutive Gene Expression| | {{Imperial/Box1|Modelling Constitutive Gene Expression| | ||
| - | A simple [[Imperial_College/Courses/Spring2008/Synthetic_Biology/Computer_Modelling_Practicals/Practical_2 | synthesis-degradation model]] is assumed for the modelling of the expression of a protein under the control of a constitutive promoter, with the same model assumed for all four [[ | + | A simple [[Imperial_College/Courses/Spring2008/Synthetic_Biology/Computer_Modelling_Practicals/Practical_2 | synthesis-degradation model]] is assumed for the modelling of the expression of a protein under the control of a constitutive promoter, with the same model assumed for all four [[Team:Imperial_College/Cloning_Strategy | promoter-RBS constructs]]. The synthesis-degradation model assumes a steady state level of mRNA. |
| - | [[Image:Eq1.png]] | + | <center>[[Image:Eq1.png]]</center> |
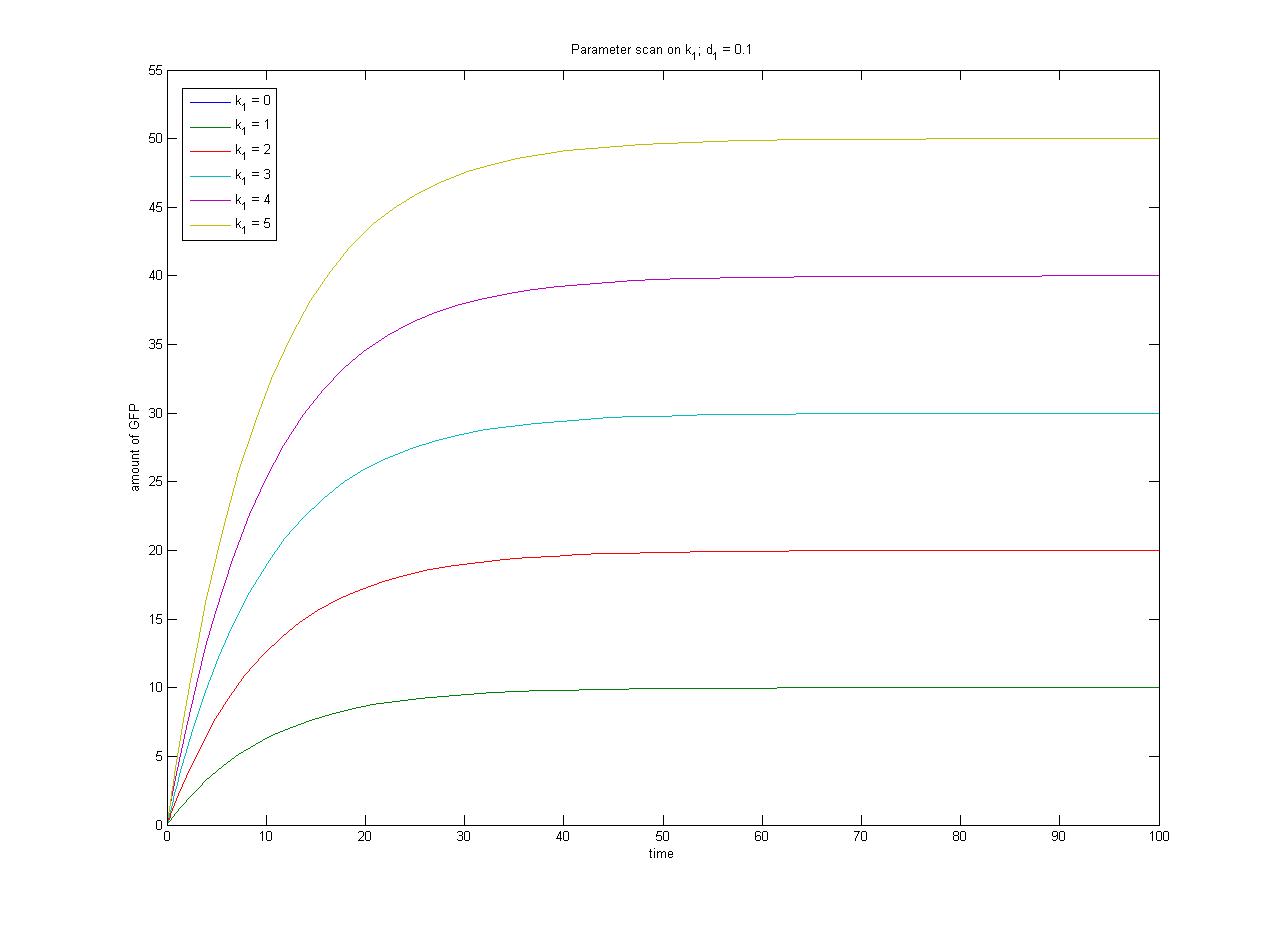
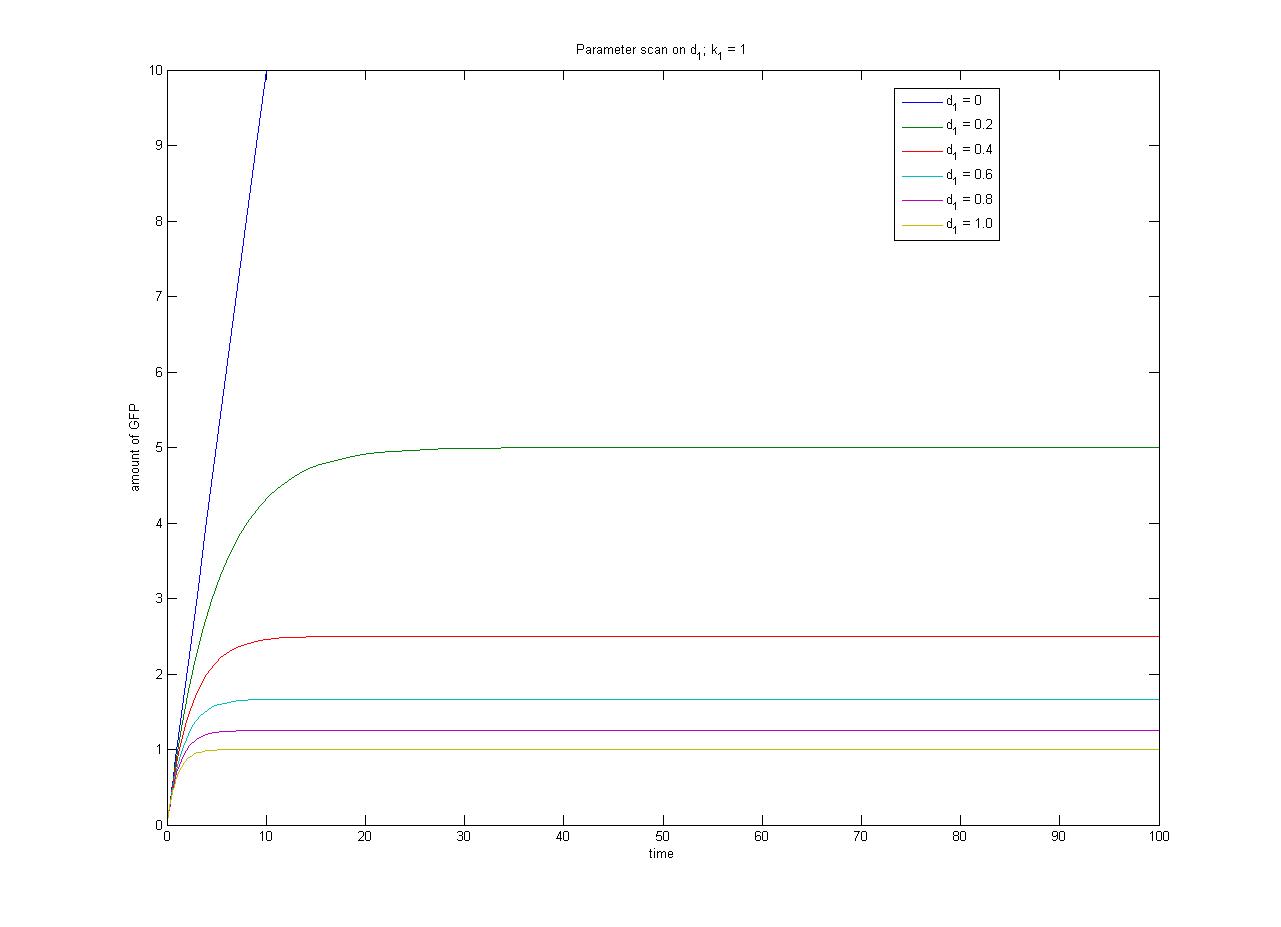
In this case, [protein] represents the concentration of GFP, k1 represents the rate of sythesis and d1 represents the degradation rate. | In this case, [protein] represents the concentration of GFP, k1 represents the rate of sythesis and d1 represents the degradation rate. | ||
| Line 17: | Line 17: | ||
We can also solve this ODE analytically. Consider the steady-state behaviour of [protein]. | We can also solve this ODE analytically. Consider the steady-state behaviour of [protein]. | ||
| - | [[Image:Eq2.png]] | + | <center>[[Image:Eq2.png]]</center> |
This relationship can be seen in the parameter scan graphs on the right. | This relationship can be seen in the parameter scan graphs on the right. | ||
| Line 34: | Line 34: | ||
{{Imperial/Box1|A simple model of inducible gene expression| | {{Imperial/Box1|A simple model of inducible gene expression| | ||
| - | |||
| - | |||
The repressor is constitutively expressed. Hence we can assume the constitutive expression model from the previous characterisation step. | The repressor is constitutively expressed. Hence we can assume the constitutive expression model from the previous characterisation step. | ||
| - | [[Image:Eq3.png]] | + | <center>[[Image:Eq3.png]]</center> |
When the inducer is added it binds reversibly to the repressor. | When the inducer is added it binds reversibly to the repressor. | ||
| - | [[Image:Eq4.png]] | + | <center>[[Image:Eq4.png]]</center> |
Repressor only binds to the promoter when it is in its unbound form, hence transcription will be a function of free repressor concentration. | Repressor only binds to the promoter when it is in its unbound form, hence transcription will be a function of free repressor concentration. | ||
| - | [[Image:Eq5.png]] | + | <center>[[Image:Eq5.png]]</center> |
And overall protein expression can be described as | And overall protein expression can be described as | ||
| - | [[Image:Eq6.png]] | + | <center>[[Image:Eq6.png]]</center> |
The [[media:ConcentrationsODE.m|ODEs]] and [[media:SimulationconcentrationsODE.m|simulation]] may be found in the Appendices section of the Dry Lab hub. | The [[media:ConcentrationsODE.m|ODEs]] and [[media:SimulationconcentrationsODE.m|simulation]] may be found in the Appendices section of the Dry Lab hub. | ||
| - | }} | + | |[[Image:Phase 2-linduced.PNG|300px]]}} |
{{Imperial/Box1|Two models of IPTG-induced expression of GFP under the control of the Plac promoter| | {{Imperial/Box1|Two models of IPTG-induced expression of GFP under the control of the Plac promoter| | ||
| Line 66: | Line 64: | ||
[[Media:InduciblePromoter_SimpleODE.m|ODEs]]<br> | [[Media:InduciblePromoter_SimpleODE.m|ODEs]]<br> | ||
[[Media:InduciblePromoterSimple.m|Simulation File]] | [[Media:InduciblePromoterSimple.m|Simulation File]] | ||
| - | |||
| - | |||
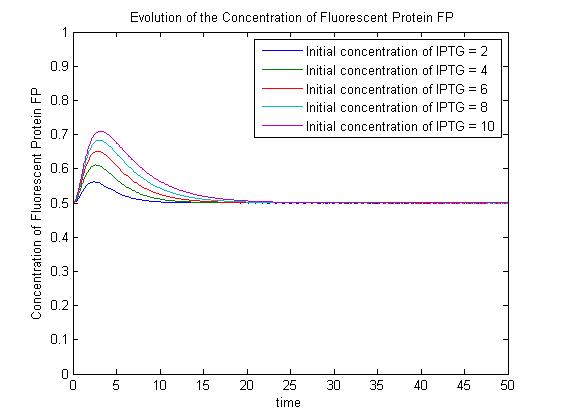
The pre-steady-state dynamic behaviour of the GFP concentration will differ with different initial concentrations of IPTG (but the steady-state behaviour will not). Hence, accuarate data collection during the pre-steady-state phase is crucial for paramater estimation. | The pre-steady-state dynamic behaviour of the GFP concentration will differ with different initial concentrations of IPTG (but the steady-state behaviour will not). Hence, accuarate data collection during the pre-steady-state phase is crucial for paramater estimation. | ||
| Line 76: | Line 72: | ||
[[Media:InduciblePromoter.m|Simulation File]] | [[Media:InduciblePromoter.m|Simulation File]] | ||
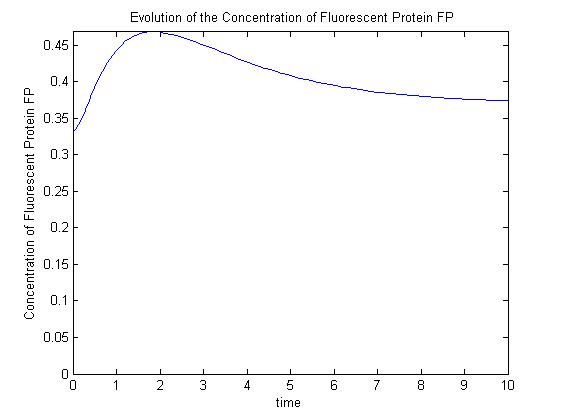
| - | [[Image:ComplexModel2.jpg|thumb|300 px|Pre-steady-state time evolution of GFP using the more sophisticated model.]] | + | |[[Image:SimpleModel.jpg|thumb|300px|Pre-steady-state time evolution of GFP using the simple model.]]<br>[[Image:ComplexModel2.jpg|thumb|300 px|Pre-steady-state time evolution of GFP using the more sophisticated model.]]}} |
| - | + | ||
| - | }} | + | |
{{Imperial/Box2|| | {{Imperial/Box2|| | ||
Revision as of 10:36, 23 October 2008
Modelling the Genetic Circuit
|
|||||||||||||||||||||||||
 "
"