Team:UC Berkeley Tools
From 2008.igem.org
(→Sponsors) |
(→Clotho Overview) |
||
| Line 16: | Line 16: | ||
==Clotho Overview== | ==Clotho Overview== | ||
[[Image:designFlow.png|thumb|700px|center|Overview of the Clotho Design Environment]] | [[Image:designFlow.png|thumb|700px|center|Overview of the Clotho Design Environment]] | ||
| + | |||
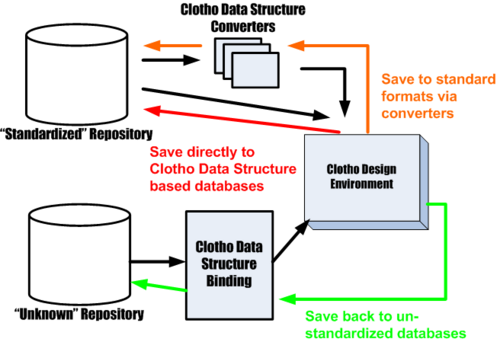
| + | Among other possible top-level explanations of Clotho, one of the simplest is the "Clotho Data Structure"-centric view. In this view, biological part data is passed back and forth between Clotho components and to files and databases outside Clotho. The possible flows of data are represented by the slew of arrows from the diagram above. | ||
| + | |||
| + | Local and standard databases vary from one lab to another, and the way to grab biological part data from one database may require a completely different (FINISH SENTENCE HERE). The Clotho Data Structure, in essence, can be viewed as the Rosetta's stone of Clotho's environment. In a nutshell, it allows developers to grab relevant information about a part. When a user wants to grab the sequence belonging to a particular Biobrick, for example, the Clotho Data Structure | ||
| + | |||
| + | Each category above merits its own explanation, as each is vitally important to the project. Perhaps a good place to start is the | ||
The Clotho design environment is centered around the ''Clotho Data Structure''. This object unifies information about a design in a standard way. This links various pieces of data related in a design to a powerful operational semantic. This semantic allows for design information to be related to databases containing part information, infer information about parts in a design, perform part look-ups in real time, and apply algorithms to designs. By taking potentially generic external information and bringing it into the Clotho Data Structure, non-standard designs can become standardized and then disseminated to a wider design community. | The Clotho design environment is centered around the ''Clotho Data Structure''. This object unifies information about a design in a standard way. This links various pieces of data related in a design to a powerful operational semantic. This semantic allows for design information to be related to databases containing part information, infer information about parts in a design, perform part look-ups in real time, and apply algorithms to designs. By taking potentially generic external information and bringing it into the Clotho Data Structure, non-standard designs can become standardized and then disseminated to a wider design community. | ||
Revision as of 17:25, 23 October 2008
With >700 available parts and >2000 defined parts in the [http://partsregistry.org/Main_Page MIT Registry of Standard Biological Parts], and doubtless more parts local to the lab, synthetic biologists are now faced with the challenge of organizing, sorting, and editing these parts from an ever-growing collection of DNA. Computational tools are becoming increasingly needed to assist biologists to do the job, and it is no secret that many tools are indeed wheeling out into the community to help serve these needs.
Clotho is a toolset designed in a platform-based-design paradigm to consolidate all these tools into one working, integrated toolbox. Clotho is based on a core-and-hub system which manages multiple connections, each connection performing tasks in some self-sufficient manner while connected to all other connections via the core. Examples of connections include sequence viewers/editors, parts database managers, sequence alignment GUIs, and much more. Computer-savvy biologists can create new connections which will then be integrated into Clotho. In this tool-consolidation and customization it is hoped that, in the future, biologists need not look further for a program that provides the answer to their needs.
Clotho Overview
Among other possible top-level explanations of Clotho, one of the simplest is the "Clotho Data Structure"-centric view. In this view, biological part data is passed back and forth between Clotho components and to files and databases outside Clotho. The possible flows of data are represented by the slew of arrows from the diagram above.
Local and standard databases vary from one lab to another, and the way to grab biological part data from one database may require a completely different (FINISH SENTENCE HERE). The Clotho Data Structure, in essence, can be viewed as the Rosetta's stone of Clotho's environment. In a nutshell, it allows developers to grab relevant information about a part. When a user wants to grab the sequence belonging to a particular Biobrick, for example, the Clotho Data Structure
Each category above merits its own explanation, as each is vitally important to the project. Perhaps a good place to start is the
The Clotho design environment is centered around the Clotho Data Structure. This object unifies information about a design in a standard way. This links various pieces of data related in a design to a powerful operational semantic. This semantic allows for design information to be related to databases containing part information, infer information about parts in a design, perform part look-ups in real time, and apply algorithms to designs. By taking potentially generic external information and bringing it into the Clotho Data Structure, non-standard designs can become standardized and then disseminated to a wider design community.
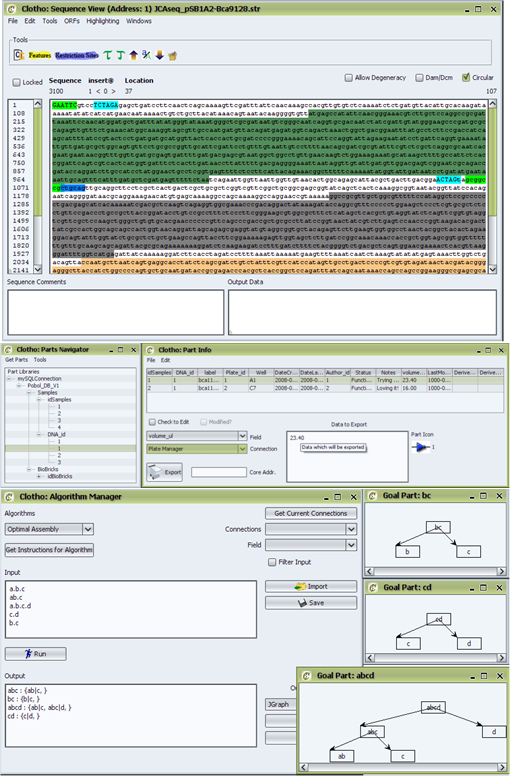
Shown in the figure are the major components in the Clotho design environment. In the center of the diagram is the Clotho Data structure...yada yada.
Clotho Database Interaction
As the synthetic biology community moves toward the standardization of parts and their repositories it is important that a design environment be agnostic when possible to the data it can begin a design with. Clotho allows for the import of both standard and non-standard data. Once the data is brought into the system it can be manipulated, analyzed, modified, and viewed. It then can either be saved back in a non-standard way as it was brought into the system initially or converted to a standardized form and distributed more widely. The figure to the right illustrates such a process.
Contact Us
For general information or to contribute: clothodevelopment AT googlegroups DOT com
For emails regarding any bugs: clothobugs AT googlegroups DOT com
For emails regarding any desired features: clothofeatures AT googlegroups DOT com
 "
"