Team:Imperial College/Genetic Circuit
From 2008.igem.org
| Line 25: | Line 25: | ||
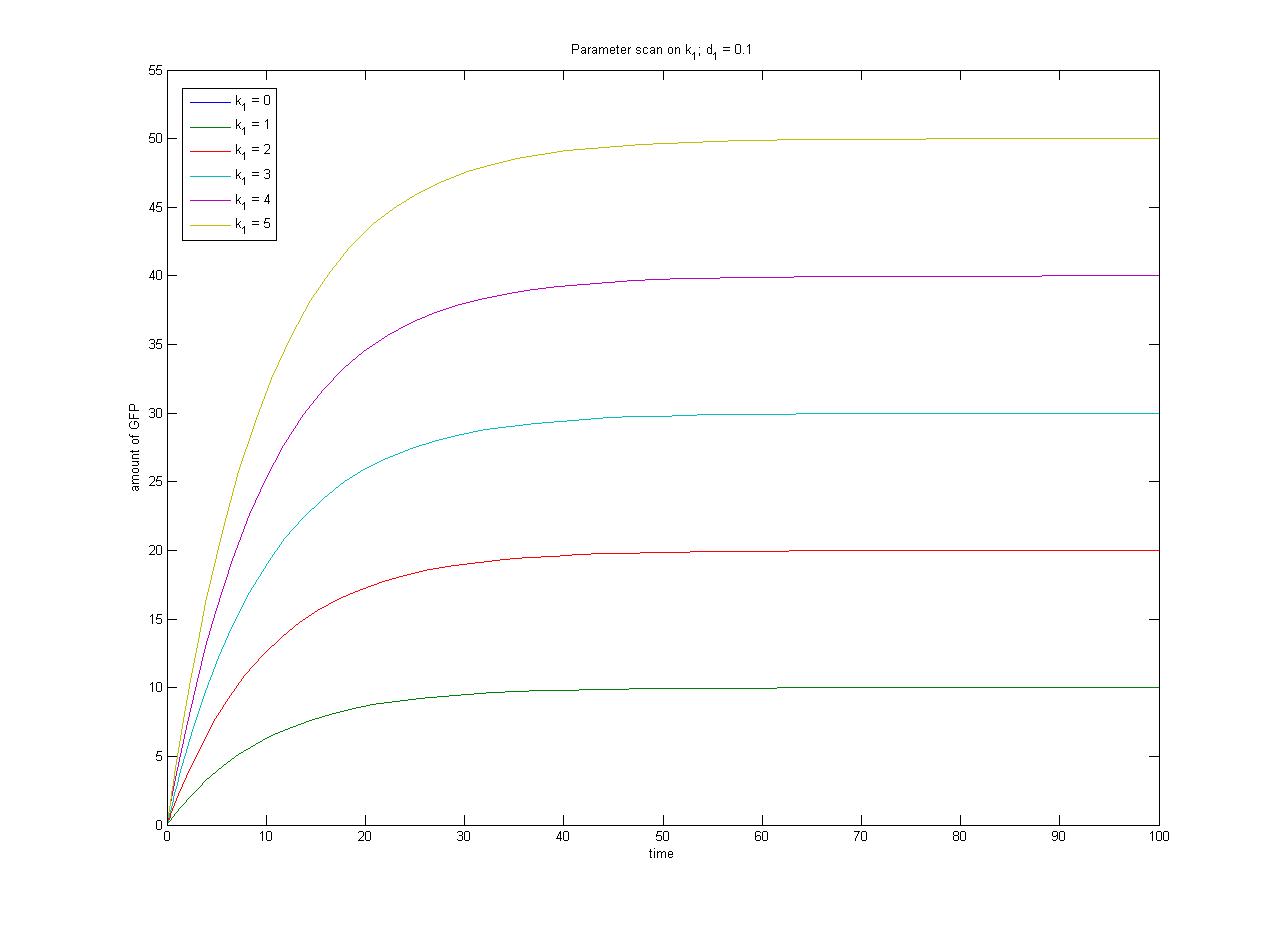
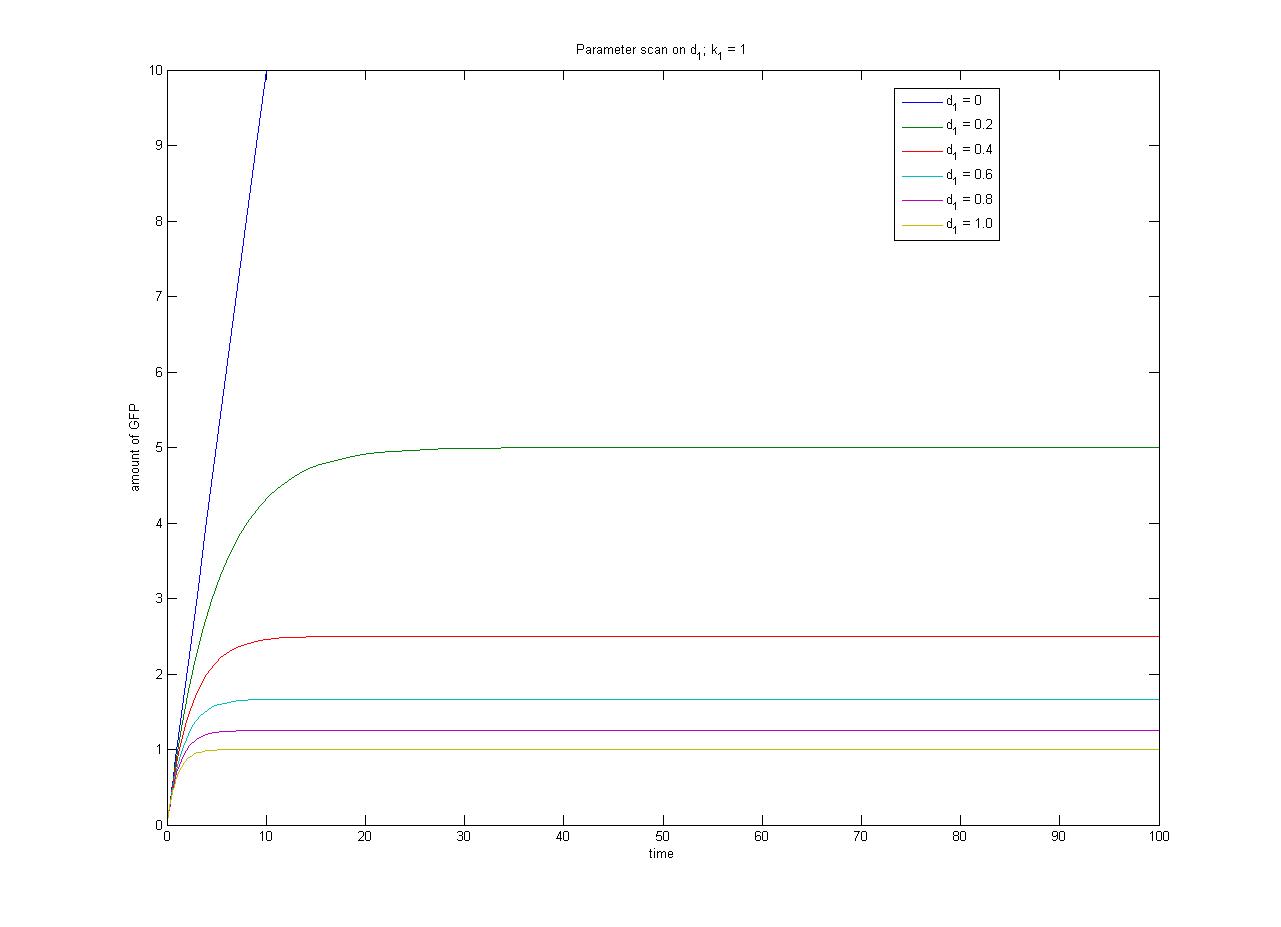
The relationship between the steady-state protein concentration and the parameters can be seen in the parameter scan graphs on the right. | The relationship between the steady-state protein concentration and the parameters can be seen in the parameter scan graphs on the right. | ||
| + | |||
| + | Note from the parameter scan graphs:<br> | ||
| + | *In the case where k1 {{Equals}} 0, no GFP is sythesised. | ||
| + | *In the case where d1 {{Equals}} 0, the concentration of protein does not reach a steady state. | ||
From the wetlab experiments it is likely that we will obtain steady-state data for each of the four promoter-RBS constructs. If we assume the same rate of degradation of GFP in each case, we can use the relative steady-state levels of GFP as a proxy for the relative rate of transcription through each promoter. This will help us with the selection of the most appropriate promoter to use for Phase 2. In order to obtain an absolute measure of transcription (as opposed to a relative measure of transcriptional strength) we require constitutive expression in terms of molecules per cell (as opposed to fluorescence in arbitrary units). | From the wetlab experiments it is likely that we will obtain steady-state data for each of the four promoter-RBS constructs. If we assume the same rate of degradation of GFP in each case, we can use the relative steady-state levels of GFP as a proxy for the relative rate of transcription through each promoter. This will help us with the selection of the most appropriate promoter to use for Phase 2. In order to obtain an absolute measure of transcription (as opposed to a relative measure of transcriptional strength) we require constitutive expression in terms of molecules per cell (as opposed to fluorescence in arbitrary units). | ||
| Line 30: | Line 34: | ||
Complementary experiments to estimate the rate of degradation of GFP - by terminating transcription - would allow us to estimate absolute rate of transcription through each promoter. | Complementary experiments to estimate the rate of degradation of GFP - by terminating transcription - would allow us to estimate absolute rate of transcription through each promoter. | ||
<br> | <br> | ||
| - | + | ||
| - | + | ||
| - | + | ||
|[[Image:Phase 1.PNG|thumb|300px|Constitutive expression of antibiotic resistance (AB) and GFP. GFP brick is part E0040, GFPmut3b. Terminator is part B0015, the double-stop.]] | |[[Image:Phase 1.PNG|thumb|300px|Constitutive expression of antibiotic resistance (AB) and GFP. GFP brick is part E0040, GFPmut3b. Terminator is part B0015, the double-stop.]] | ||
Revision as of 18:27, 27 October 2008
Modelling the Genetic Circuit
--Mabult 14:44, 27 October 2008 (UTC) Cut this part out and move it into Pridence's section (simple repression Model)
|
|||||||||||||||||||||||||
 "
"