Team:Calgary Software/Project
From 2008.igem.org
(→Evolutionary Algorithm) |
(→Data Retrieval and Storage) |
||
| Line 40: | Line 40: | ||
== [[Data Retrieval and Storage]] == | == [[Data Retrieval and Storage]] == | ||
| - | [[Image:Perl logo.PNG|thumb| | + | [[Image:Perl logo.PNG|thumb|right|150px]] |
To improve EvoGEM as a model, organic compounds and biochemical reactions needed to be examined. Using a set of PERL scripts, key pieces of data is retrieved from the source information of the registry. For every part, the type must be known, its function, whether it codes for a protein, and how well the parts work. If a part codes for a protein, its DNA sequence is retrieved. The amino acid sequence is found (using the BLAST algorithm) from UniProt, which is a large database of proteins. If the protein has an associated prosthetic or a biochemical reaction, this is also examined. From there, additional information is retrieved from ChemSpider - a chemical database - to find the data that characterizes any compounds that are involved with that particular protein. Finally, the data is stored in a data base during run-time of EvoGEM. Refer to the [[Data Retrieval and Storage]] page for further details. | To improve EvoGEM as a model, organic compounds and biochemical reactions needed to be examined. Using a set of PERL scripts, key pieces of data is retrieved from the source information of the registry. For every part, the type must be known, its function, whether it codes for a protein, and how well the parts work. If a part codes for a protein, its DNA sequence is retrieved. The amino acid sequence is found (using the BLAST algorithm) from UniProt, which is a large database of proteins. If the protein has an associated prosthetic or a biochemical reaction, this is also examined. From there, additional information is retrieved from ChemSpider - a chemical database - to find the data that characterizes any compounds that are involved with that particular protein. Finally, the data is stored in a data base during run-time of EvoGEM. Refer to the [[Data Retrieval and Storage]] page for further details. | ||
<br style="clear:both"/> | <br style="clear:both"/> | ||
Revision as of 21:44, 28 October 2008

|
| Home | The Team | The Project | Modeling | Notebook |
|---|
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
Contents |
Introduction
EvoGEM was briefly presented during the 2007 iGEM jamboree and has sparked quite a lot of interest amongst the different teams. This summer, our team plans to further develop the fitness function EvoGEM employs, introduce more complex pattern recognition, and test the system under a much larger search space than before. The final goal is to produce a system sophisticated enough to rebuild working designs from previous years' teams' projects, as well as intelligent enough to simulate successes and failures of working and non-working systems, respectively. The main focus of this project is to build perl scripts that will support EvoGEM's requirements of a flat file registry, create an Objective-C based graphical user interface (GUI) in order to make the software-user interaction easy for any potential users, develop the EvoGEM code to include the behaviors specified before, and create a simulation of the processes in the cell such as transcription and translation.
Evolutionary Algorithm
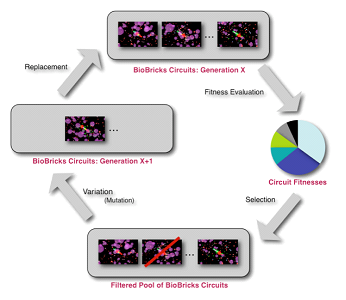
Evolution involves the changes of inherited traits in a population from successive generations, one to another. Genetic information is carried through each generation and certain characteristics are expressed. Mutation enables manipulation of these traits as well as genetic recombination. Evolution is the result from the heritable traits becoming more prevalent or rare.
Agent-based modeling is a computational method of replicating the behavior and interaction of individuals within a network such that their overall effect on the system can be observed. This involves many different aspects, including game theory, evolutionary programming, complex, systems, and emergence. Multiple agents are simulated throughout an environment to emulate and hypothesize the actions of complex phenomena. Refer to Evolutionary Algorithm page for more details.
Data Retrieval and Storage
To improve EvoGEM as a model, organic compounds and biochemical reactions needed to be examined. Using a set of PERL scripts, key pieces of data is retrieved from the source information of the registry. For every part, the type must be known, its function, whether it codes for a protein, and how well the parts work. If a part codes for a protein, its DNA sequence is retrieved. The amino acid sequence is found (using the BLAST algorithm) from UniProt, which is a large database of proteins. If the protein has an associated prosthetic or a biochemical reaction, this is also examined. From there, additional information is retrieved from ChemSpider - a chemical database - to find the data that characterizes any compounds that are involved with that particular protein. Finally, the data is stored in a data base during run-time of EvoGEM. Refer to the Data Retrieval and Storage page for further details.
Modeling
Graphical User Interface
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
| Home | The Team | The Project | Modeling | Notebook |
|---|
 "
"