Team:PennState/diauxie/progress
From 2008.igem.org
| Line 239: | Line 239: | ||
<div class="gallerytext"><p>W3110 fluoresence</p></div> | <div class="gallerytext"><p>W3110 fluoresence</p></div> | ||
</div> | </div> | ||
| + | |||
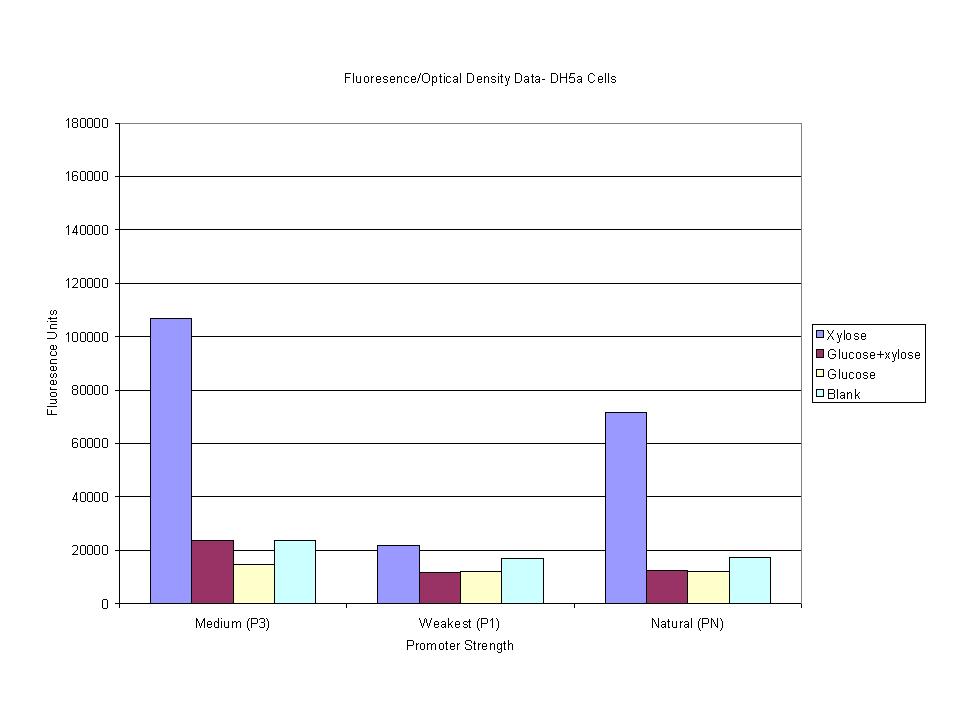
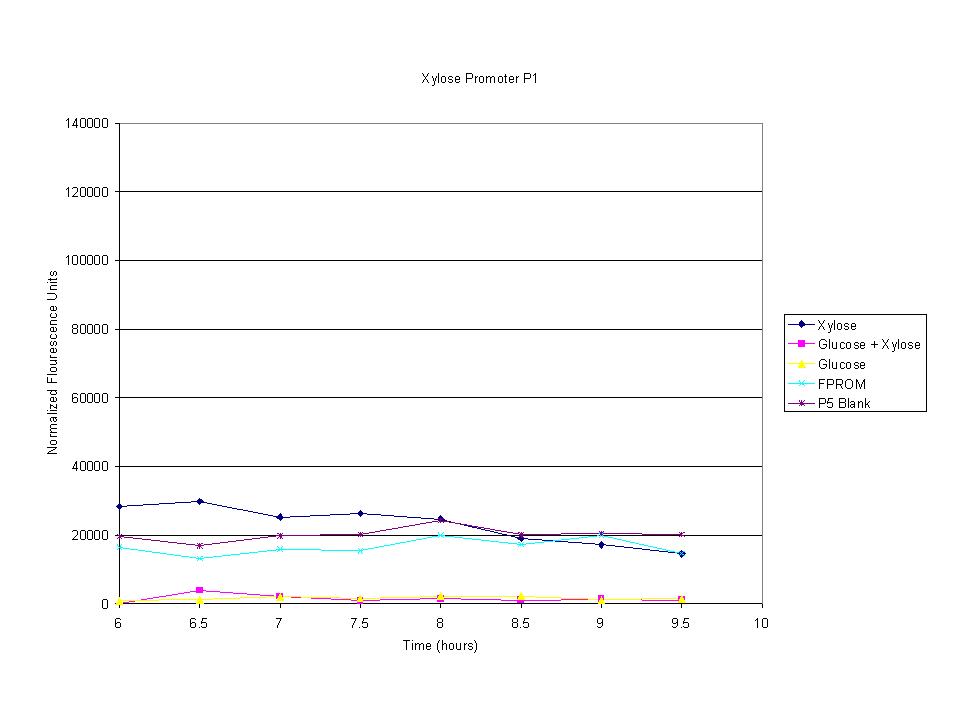
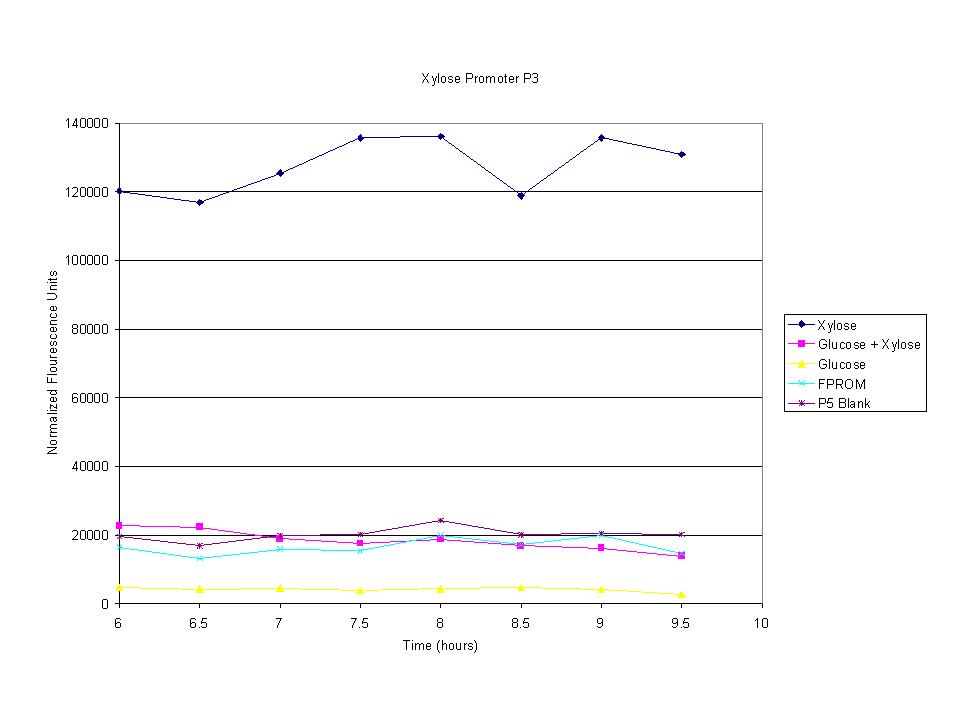
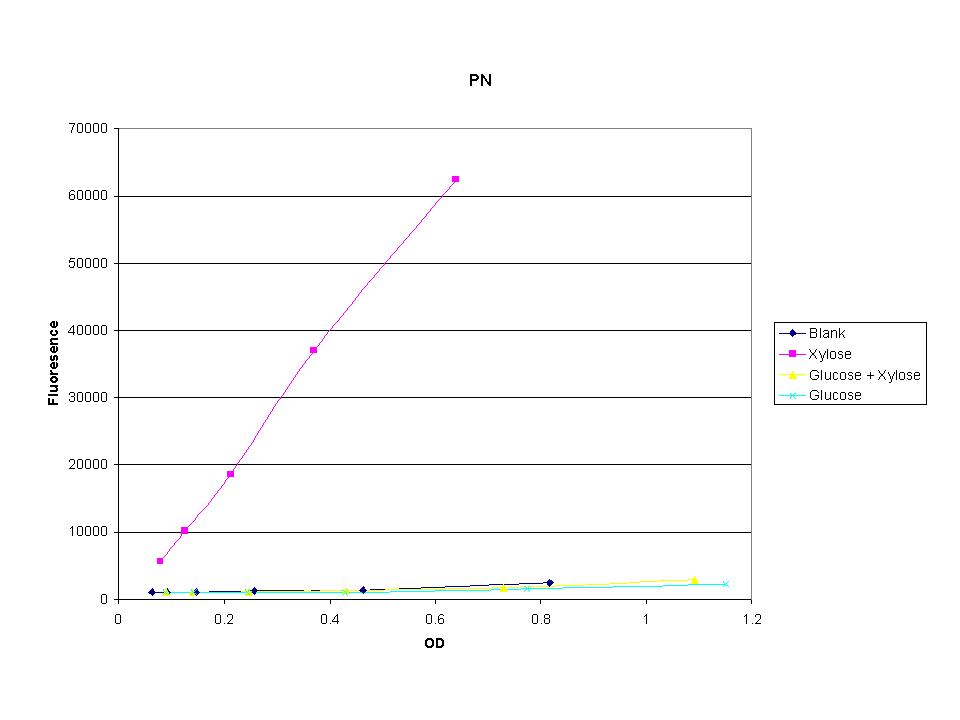
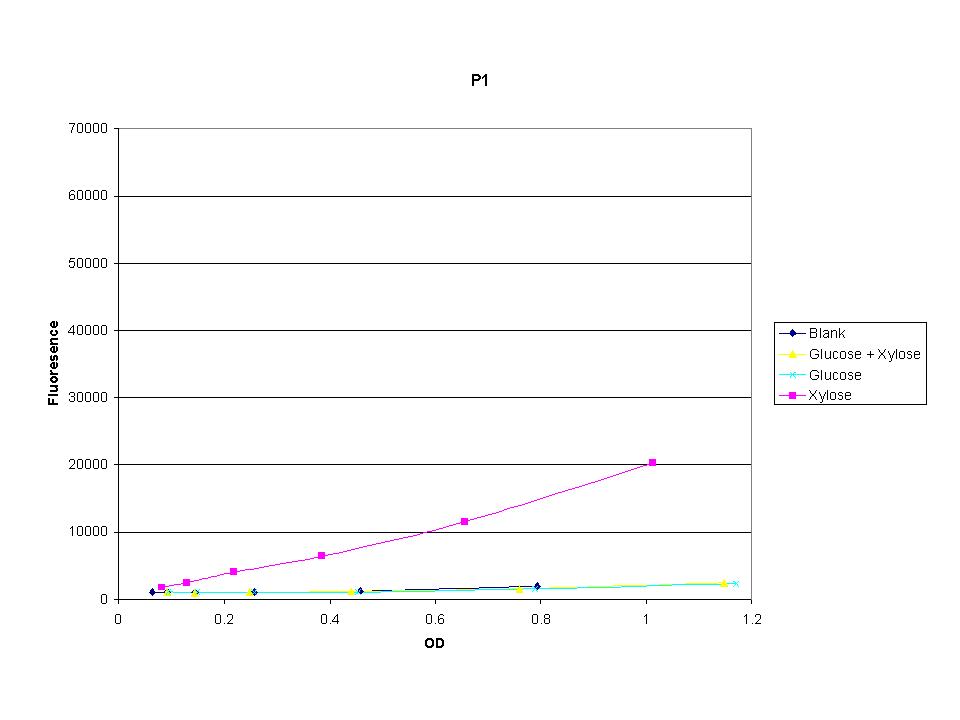
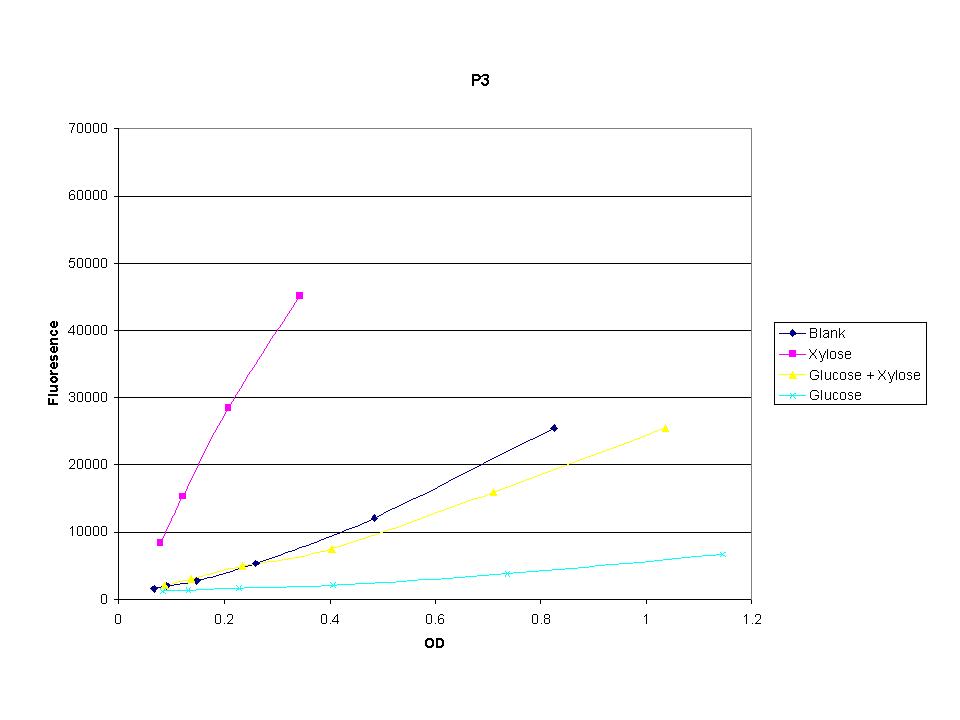
| + | <p>These graphs show the normalized fluoresence strength for PN, P1, and P3 xylose promoters induced with xylose, glucose, and a mixture. The W3110 cells have xylE and xylG knocked out while DH5a contains the natural E. coli chromosome. This data shows that there is little effect on the fluorescence intensity using strains with xylE and xylG sequences deleted. Our next step is to transform these promoters into E. coli cells with deleted xylose metabolism and transporters. </p> | ||
</div> | </div> | ||
Revision as of 15:59, 29 October 2008

| Home | The Team | The Project | Parts | Notebook |
Diauxie EliminationNHR Biosensors
|
Progress & Results
We have completed our first set of cloning phases and are conducting our first round of testing and modification. Each test construct (promoter + GFP) has been cloned into pSB1A2 and transformed into DH5α, W3110 ∆xylB-G, and W3110 ∆xylB-R. Preliminary induction studies were run to find the optimal induction time and to see in what range OD and fluorescence were linearly related. Test Construct![[Test Consturct]](https://static.igem.org/mediawiki/2008/1/1b/Test_construct.JPG)
|
 "
"