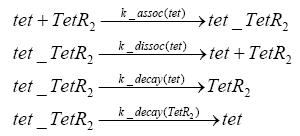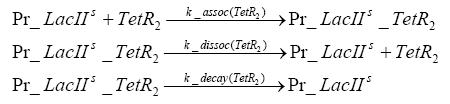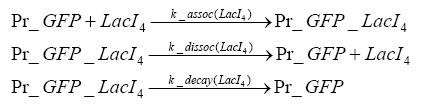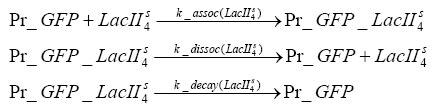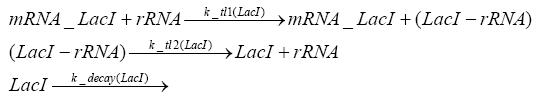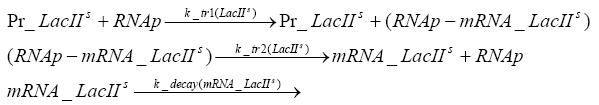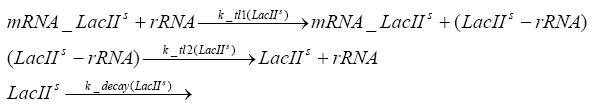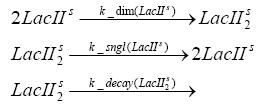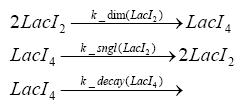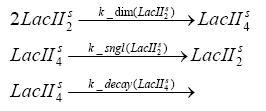Team:ETH Zurich/Modeling/Switch Circuit
From 2008.igem.org
(→Parameters) |
(→Parameters) |
||
| Line 224: | Line 224: | ||
In this section you can find all the parameters used in the simulation.<br> | In this section you can find all the parameters used in the simulation.<br> | ||
| - | Because many of the in vivo rates of the biochemical reactions we simulated are unknown | + | Because many of the in vivo rates of the biochemical reactions we simulated are unknown or could not be found in the literature, the kinetic parameters were mainly obtained from estimates [https://2008.igem.org/Team:ETH_Zurich/Modeling/Switch_Circuit#References (2)] based on the values found in the supporting text to [https://2008.igem.org/Team:ETH_Zurich/Modeling/Switch_Circuit#References (1)].<br> |
{| border="1" | {| border="1" | ||
Revision as of 01:12, 30 October 2008
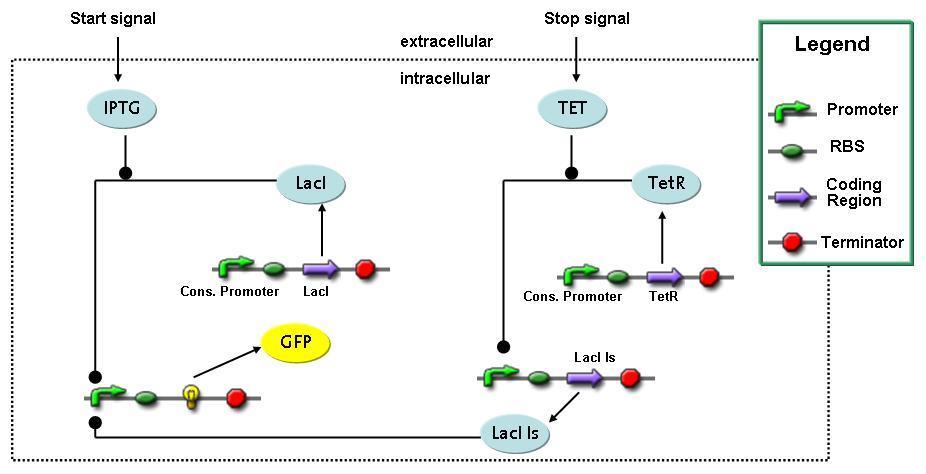
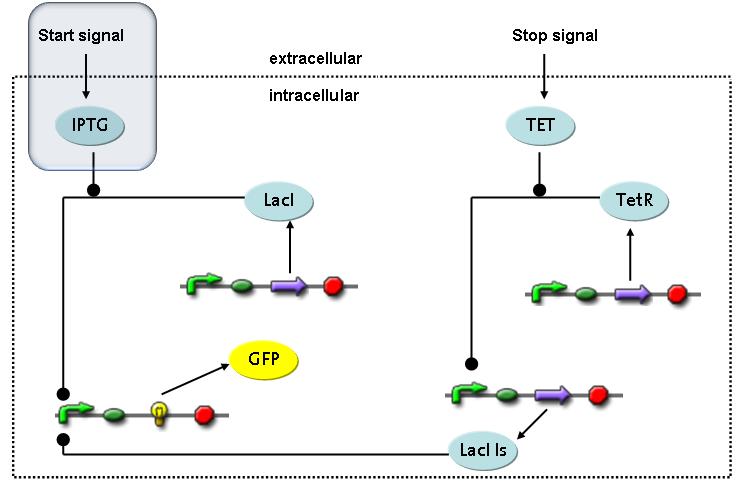
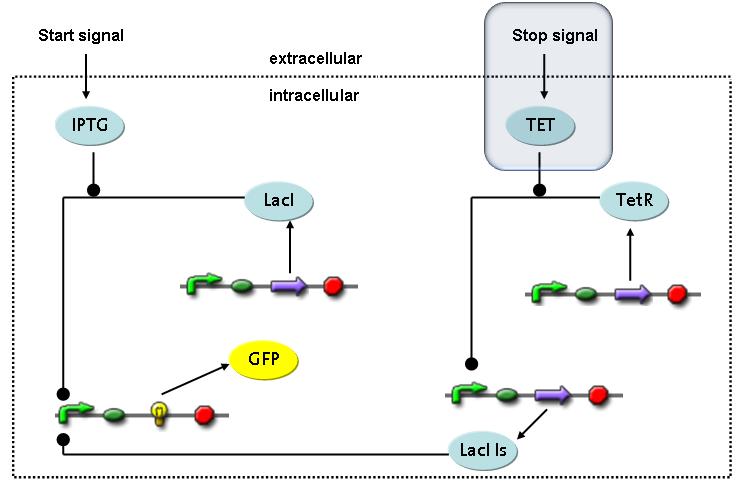
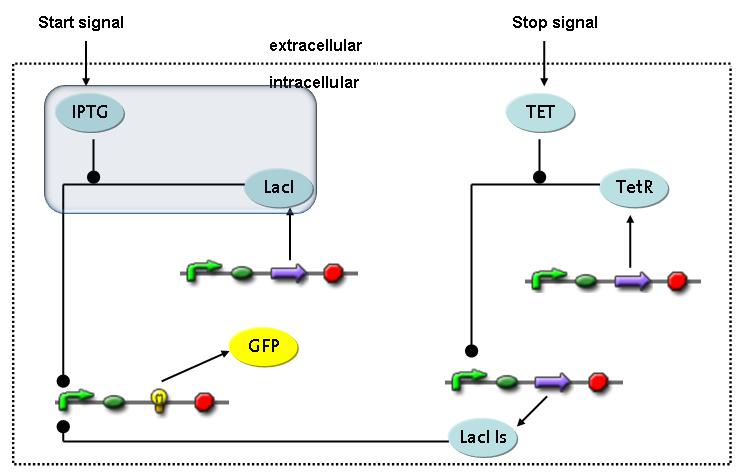
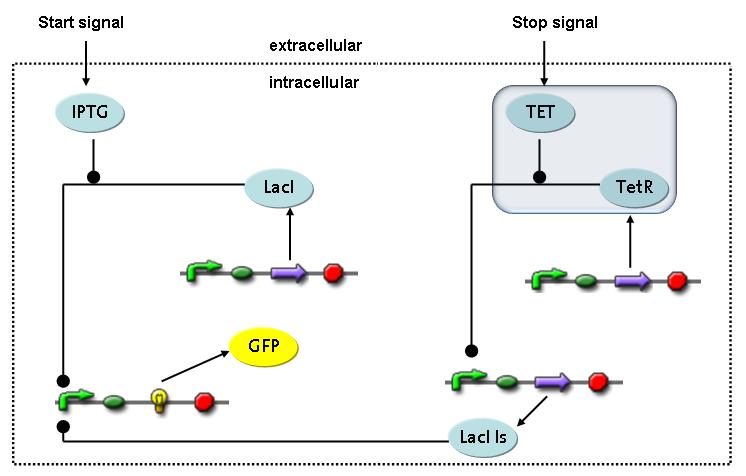
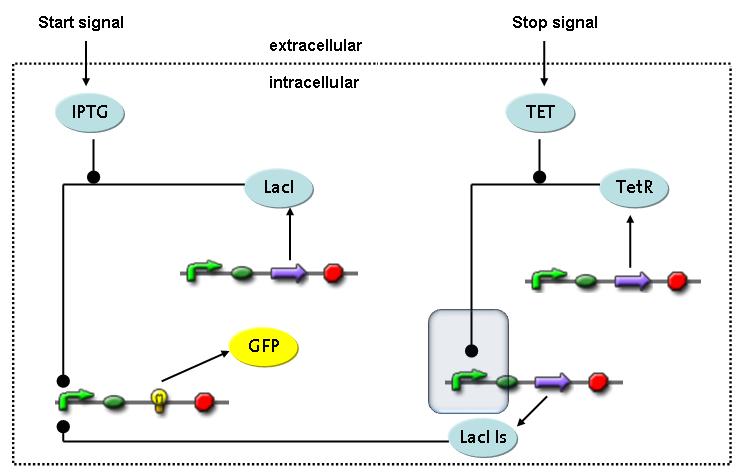
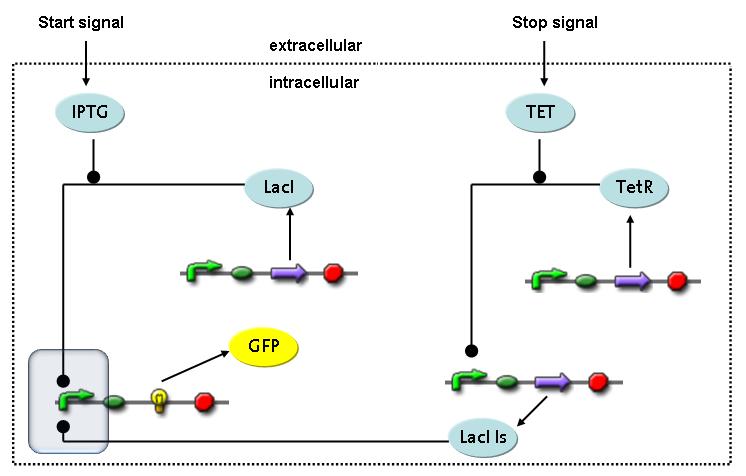
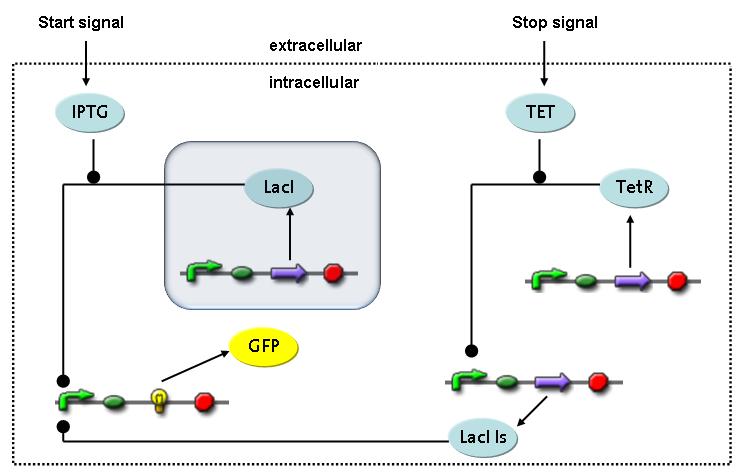
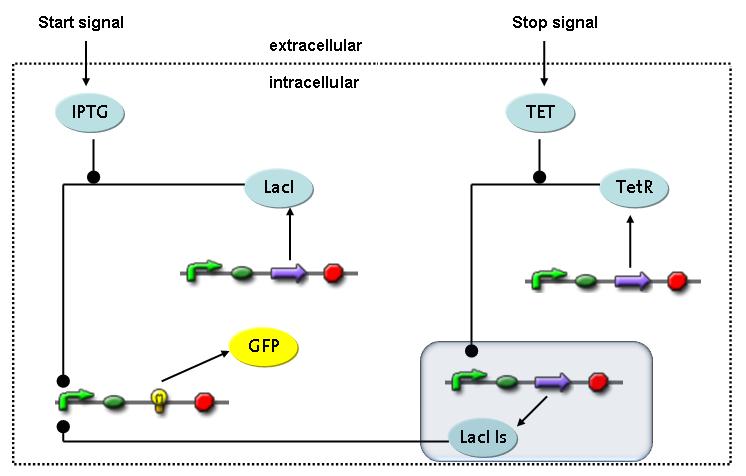
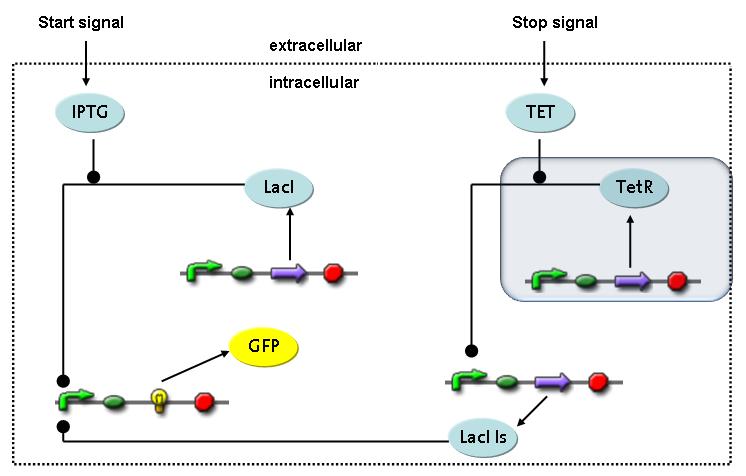
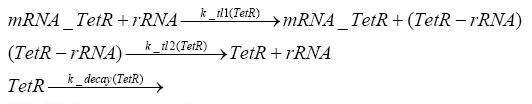
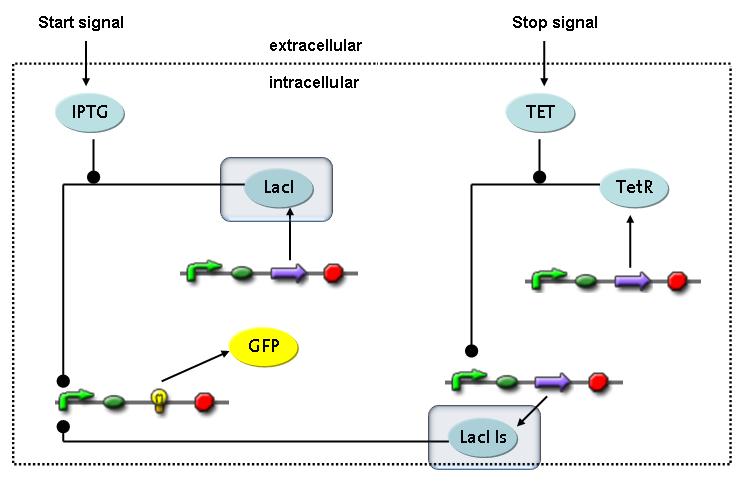
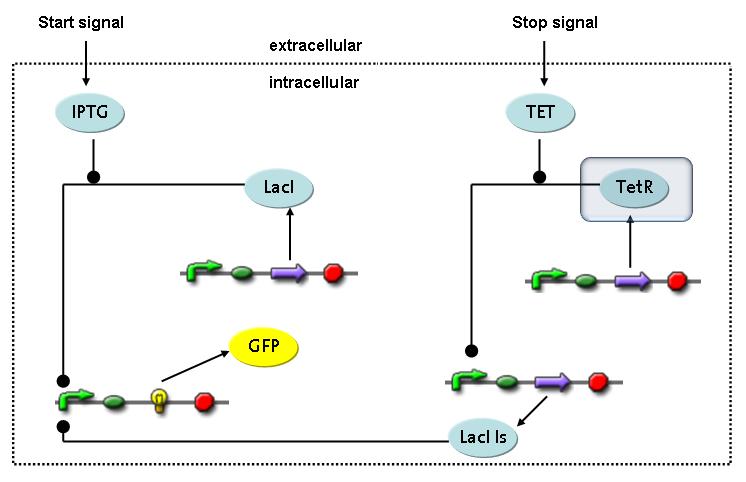
Switch CircuitThe designed switching circuit is driven by two input signals – a start signal initiates the synthesis of a specific protein and a terminating signal switches gene expression off. The goal of this system is to control expression of a restriction enzymes in order to delete genome fragments in vivo.
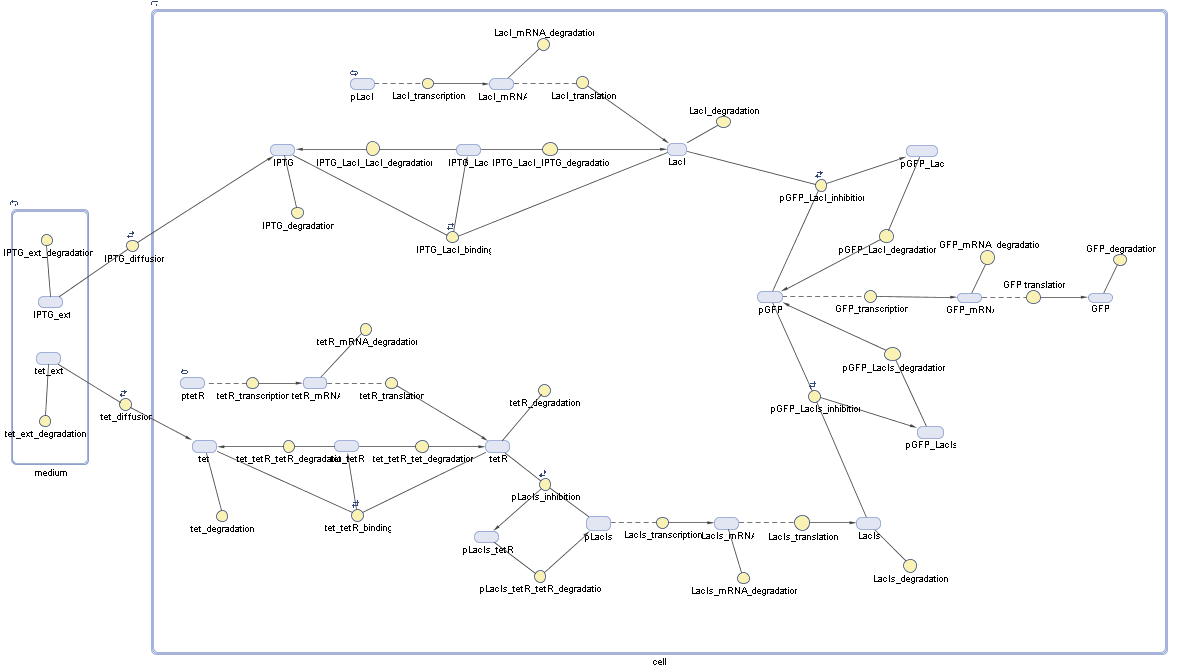
In order to do some preliminary experiments, the restriction enzyme has been substituted by a fluorescent protein. The detailed mechanism is described here. It follows a summarized version of it's mode of operation: This switching circuit is described by a set of 64 chemical reactions and 41 molecular species. In order to do a computational analysis of the circuit, this model has been simplified and implemented in MATLAB. Then the system has been simulated using ODE and stochastic solvers. Implementation and SimulationImplementationFor the implementation, different steps that do not have significant effects on the aspired results have been neglected for the sake of simplicity. The transcriptional step involving the RNA-Polymerase and the translational step involving the ribosomes have not been taken into account. Furthermore, the effects of the dimerization of TetR as well as the impacts of dimerization and tetramerization of LacI and LacIIs (4) have not been considered in the final implementation. This simplified model still comprises more than 20 different species and over 30 kinetic reactions and was implemented by using the SimBiology Toolbox in MATLAB. We performed deterministic and stochastic simulations based on Mass-Action-Kinetics. The stochastic simulations turned out to be computationally very exhaustive but generated no further significant information compared to the deterministic simulations. Simulation Results
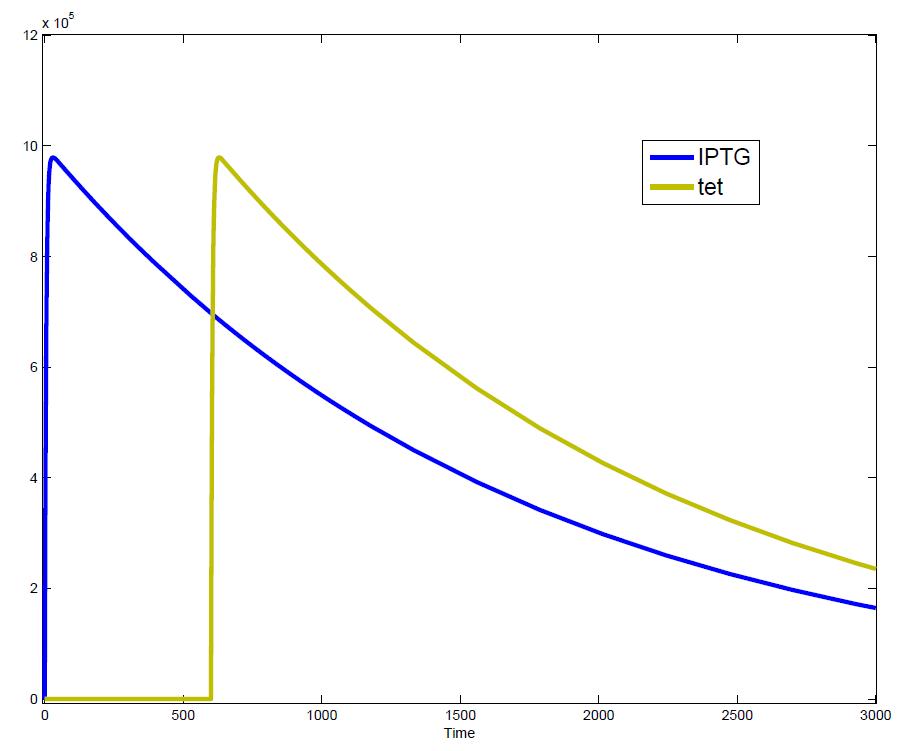
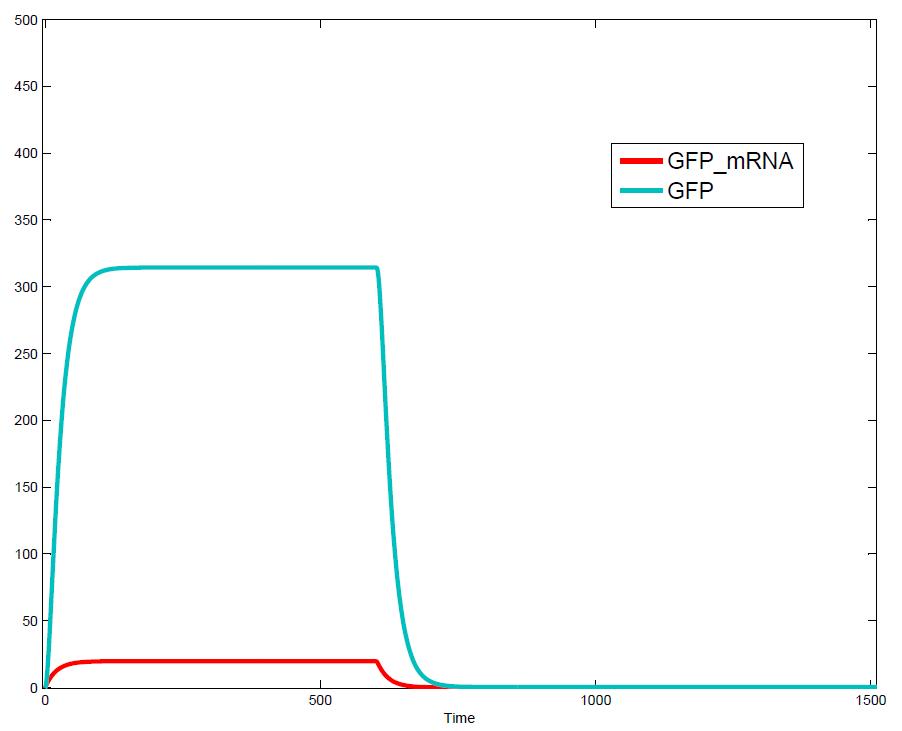
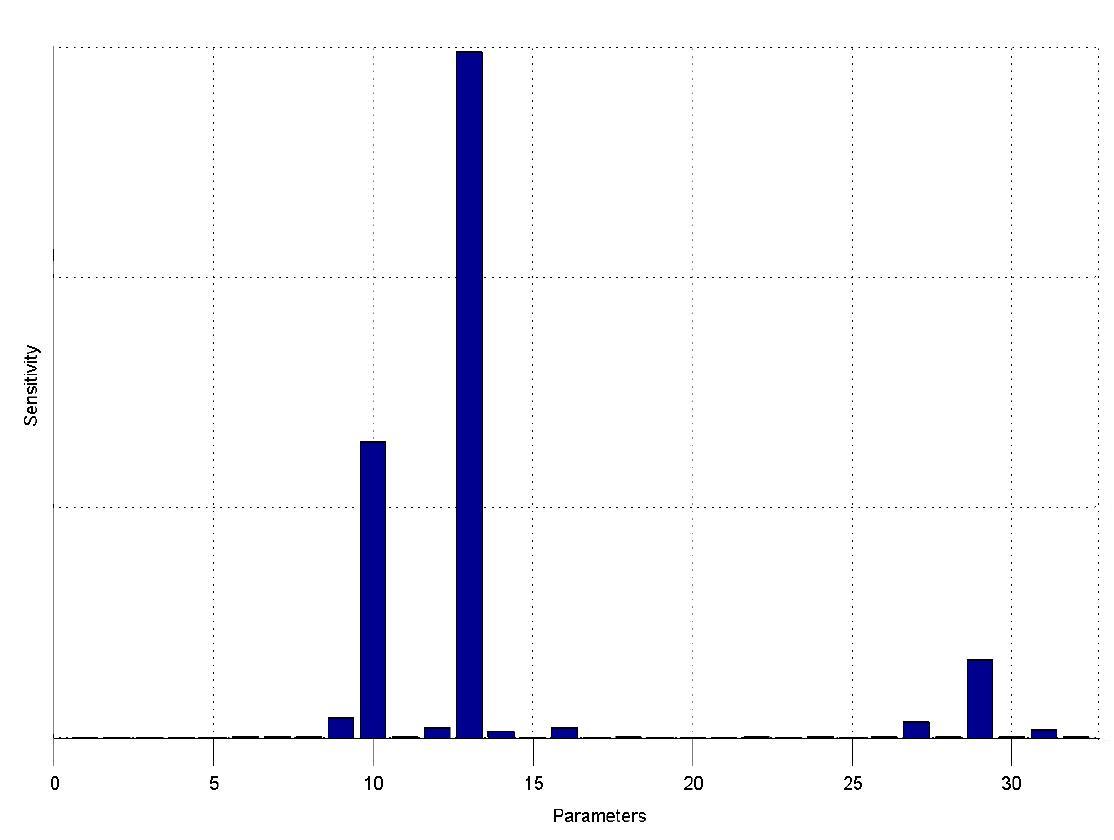
The simulations show that our system actually should create a nice pulse-shaped expression of the fluorescent protein (GFP). This expression can be initiated by inducing with IPTG and stopped by subsequent addition of tet into the medium. By tagging the protein it will be degraded much faster by the Clp protease, so that the overall concentration is bounded and, after activating the stop-signal, the remaining proteins disappear quickly. Another fact that the simulations showed is that in order to get one single pulse the tet-concentration inside, the medium must not reach zero before all the IPTG is degraded too. Otherwise there would still be IPTG in the system inhibiting the binding of LacI to the GFP-promoter and leading to an unwanted expression of our protein of interest as the LacIIs degrades. One way to overcome this problem is by simply inducing with a much higher quantity of tet than IPTG, so that it simply takes longer for it to completely degrade or being washed away. Still another way would be to wait a bit longer after the induction with IPTG so that it has already partly vanished until switching off the GFP-expression with tet. Sensitivity AnalysisWe define the sensitivity as the change of the production of the desired fluorescence protein - which is the output of our system - depending on the change of the parameters.
The sensitivity analysis shows, that the concentration of the fluorescent protein strongly depends on its decay rate (parameter 13) the decay rate of its mRNA (parameter 10) and of course the transcription and translation rates of the protein (parameters 29 and 27), which is no surprise.
We can also see that the decay rate of LacIIs (parameter 9) and the transcription rate of LacIIs (parameter 31) have an influence on the expression of the fluorescent protein.
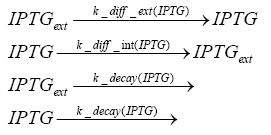
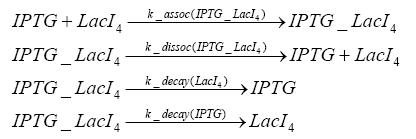
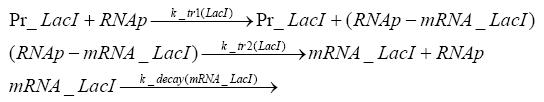
Detailed Modeldiffusion of IPTGIn order to switch on the circuit, we induce with IPTG. When IPTG is added into the medium it diffuses reversibly between the medium and the cells, where it is slowly degraded. In a chemostat extracellular IPTG is washed away. diffusion of tetThe second inducer which is used in our system is tet. This one also ddiffuses reversibly between the medium and the cells, where it is slowly degraded and is washed away in the medium. binding of IPTG to LacIThe first inducer IPTG can bind to the tetramerized LacI (4). binding of tet to TetRThe second inducer tet can bind to TetR. binding of TetR to LacIIs-promoterTetR is constitutively expressed and binds to the LacIIs promoter, inhibiting its expression. binding of LacI and LacIIs to GFP-promoterLacI which is constitutively expressed and LacIIs which is under the control of a tet repressor can bind both to the GFP promotor. transcription and translation of LacI
transcription and translation of LacIIs
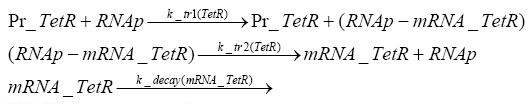
transcription and translation of TetR
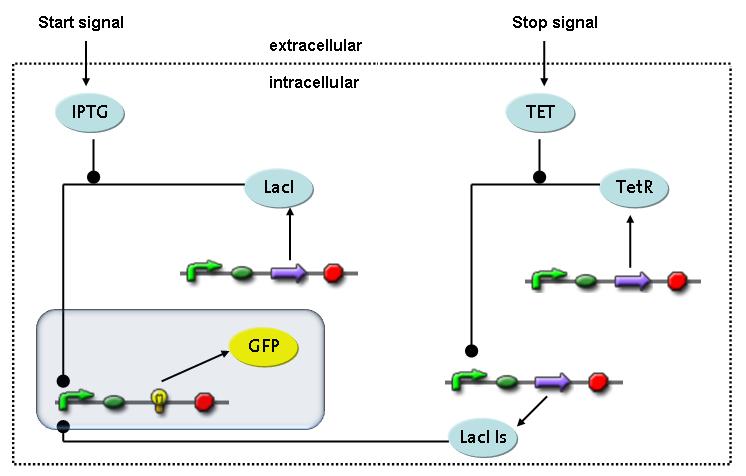
transcription and translation of GFP
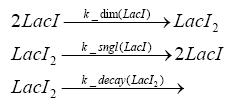
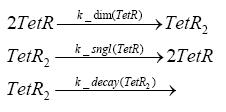
dimerization and tetramerization of LacI and LacIIsdimerization of tetRParametersIn this section you can find all the parameters used in the simulation. Because many of the in vivo rates of the biochemical reactions we simulated are unknown or could not be found in the literature, the kinetic parameters were mainly obtained from estimates (2) based on the values found in the supporting text to (1).
[*] the degradation constants of the two inducers are bigger outside the cell, the effect that they are washed away in the chemostat is taken into account in those parameters References(1) "Spatiotemporal control of gene expression with pulse-generating networks", Basu et al., PNAS, 2004 (2) "Genetic circuit building blocks for cellular computation, communications, and signal processing", Weiss et al., Natural Computing, 2003 (3) "Predicting stochastic gene expression dynamics in single cells", Mettetal et al., PNAS, 2006 (4) "Engineered gene circuits", Hasty et al., Nature, 2002 |
 "
"