Team:Chiba/Project
From 2008.igem.org
(→Delay Switching by Tuning Components) |
(→Delay Switching by Tuning Components) |
||
| Line 90: | Line 90: | ||
'''[[Team:Chiba/Experiments:LuxR_mutant|Lux Mutants]]''' | '''[[Team:Chiba/Experiments:LuxR_mutant|Lux Mutants]]''' | ||
| width="25%" valign="top" align="center"| | width="25%" valign="top" align="center"| | | width="25%" valign="top" align="center"| | width="25%" valign="top" align="center"| | ||
| - | | width="25%" valign="top" align="center"| | + | | width="25%" valign="top" align="center"| | width="25%" valign="top" align="center"| |
| - | | width="25%" valign="top" align="center"| | + | |
|} | |} | ||
Revision as of 02:04, 30 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
Abstract
E.coli time manager
We are constructing delay switches to control/preset the timing of target gene expression. Our project uses two classes of bacteria: senders and receivers. Senders produce signaling molecules, and receivers are activated only after a particular concentration of this molecule is reached. The combinatorial use of senders/receivers allows us to make a‘switching consortium’which activates different genes at the preset times.
As signaling molecules, we utilize molecules associated with Quorum sensing, a phenomenon that allows bacteria to communicate with each other. Although different quorum sensing species have slightly different signaling molecules, these molecules are not completely specific to their hosts and cross-species reactivity is observed (1),(2). Communication using non-endogenous molecules is less sensitive than the original, and requires a higher signal concentration to take effect. This reduced sensitivity results in the slower activation of receivers, thus creating a system in which different receivers are activated after different amounts of time following signaling molecule release.
Introduction
Many electronic devices we use in our daily lives have the ability to keep track of time. For example, a VCR is able to record a TV program at a pre-set time, and a microwave automatically stops heating after a set amount of time. automatically stop heating when the right time comes. Using these temporal pre-programming functions, we have been liberated from either staying up late to watch a European soccer game or from worrying about our popcorn being burned black while yelling and shouting to the match we have videotaped. In this way, the timer function has revolutionized our lifestyle.
We thought the same applies to the biotechnology; we would like to freely implement the 'timer switches” to various biological functions, preferably both independently and in parallel format. These “functions” include sensors, synthesizers, or degraders of bioactive compounds/ materials, transportation and secretion machineries, communications, getting/ sticking together, proliferation and cell death. If successful, we will be able to program exceedingly more complex complex behaviors in cellular systems.
a while, the message is gone and cannot be retrieved.
Project Design
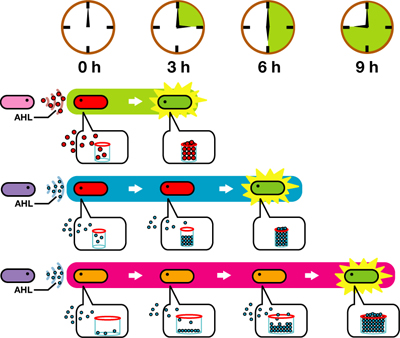
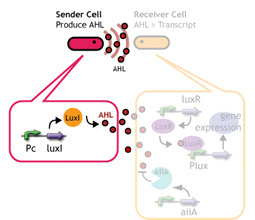
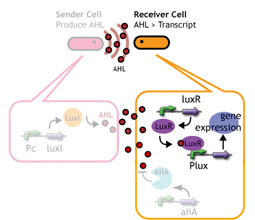
We designed a "switching consortium" that works like a water clock ([http://en.wikipedia.org/wiki/Water_clock Water clock-wikipedia.en]). Here is how it works (Fig X).
1. Sender cells slowly generate signal molecules AT A CONSTANT RATE. The signal molecules are non-degradable so they get accumulated (linearly) over time.
2. Receivers detect the signal molecules and then activate the genetic switch to the on state, but only when the signal concentration reaches the switching threshold of a receiver. Receivers are activated by different signaling molecules at different rates. In this way, the entire system behaves like a delay switch sequence.
3. Either by changing the receiver sensitivity or rate of signal accumulation, one can freely control the delay length of the individual switches. Using switches with various times-of-delay, one can sequentially activate many different cellular functions.
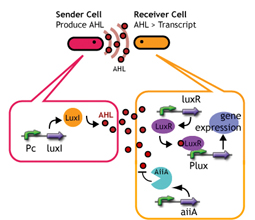
Signaling System
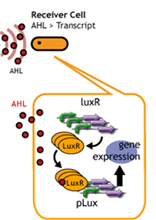
In this project, we use acylated homoserine lactones (AHLs), signaling molecules used for [http://en.wikipedia.org/wiki/Quorum_sensing quorum sensing] in gram negative bacteria. Senders express LuxI or similar enzymes, which catalyze the production of AHLs, under the control of a constitutive (Tet) promoter. Each cell thus generates AHL more or less at a constant rate. AHL can freely permeate cell membranes and are detected by neighboring cells. Receivers constitutively express LuxR proteins (or a similar ortholog), the protein that detects AHL concentrations. When AHLs bind LuxR proteins, the AHL-LuxR complex activates the Lux promoter. The threshold [AHL] at which switching occurs is determined by the affinity of AHL for the particulr LuxR ortholog.(3),(4). (more about quorum sensing)
Constructing A Delay Switch, three ways:
- Speaker whispering
- Desensitize Receiver
- jamming by AiiA (decomposer)
List of Experiments
Delay Switching by Tuning Components
| | width="25%" valign="top" align="center"| | | width="25%" valign="top" align="center"| |
Delay Switching by CrossTalk(7)
Miscs
Conclusion
When we used the cross-talk strategy using 3OC12HSL instead of 3OC6HSL, the delayed switch worked 2 hours after the original switch worked. Futher variation of delayed switches will be realized:
- Combination of senders and receivers through cross-talk.
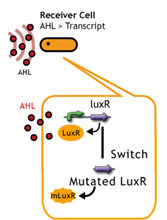
- LuxR mutation(receiver).
- Make a gene circuit of positive feed back loop connected quorum sensing.(8),(9)
References
- [http://www3.interscience.wiley.com/journal/119124142/abstract M.K Winson et al.:Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing.FEMS Microbiology Letters 163 (1998) 185-192]
- [http://partsregistry.org/Part:BBa_F2620:Specificity BBa_F2620:Specificity]
- [http://www.pnas.org/content/100/suppl.2/14549.full Michiko E. Taga. Bonnie L.Bassler.:Chemical communication among bacteria.PNAS.November 25, 2003,100.suppl.2]
- [http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.micro.55.1.165?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dncbi.nlm.nih.gov Melissa B. Miller and Bonnie L. Bassler.:QUORUM SENSING IN BACTERIA.annurev.micro.55.1.165.2001]
- [http://authors.library.caltech.edu/5553/ C. H. Collins.et al.:Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones.Mol.Microbiol.2005.55(3).712–723]
- [http://mic.sgmjournals.org/cgi/content/abstract/151/11/3589 B. Koch. et al.:The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors.Microbiology 151 (2005),3589-3602]
- [http://mic.sgmjournals.org/cgi/content/full/153/12/3923 Paul Williams :Quorum sensing, communication and cross-kingdom signalling in the bacterial world.Microbiology 153 (2007), 3923-3938]
- [http://pubs.acs.org/cgi-bin/abstract.cgi/acbcct/2006/1/i11/abs/cb6004245.html D. J. Sayut et al.:Construction and Engineering of Positive Feedback Loops.ACS Chemical Biology.1.No.11.(2006)]
- [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WBK-4PRHJ6K-2&_user=136872&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_version=1&_urlVersion=0&_userid=136872&md5=3ed7ffab9f831d102f5e99353843c080 D. J. Sayut et al.:Noise and kinetics of LuxR positive feedback loops.Biochem. Biophys. Res. Commun.363(3),2007,667-673.]
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
 "
"