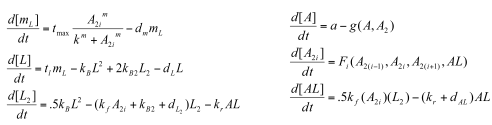From 2008.igem.org
(Difference between revisions)
Revision as of 02:45, 30 October 2008
|

|
|
Sequestillator Model 2: A More Complicated Model
While Model 1 gave us some important results (mainly: need a low Kd for oscillations to occur) , we decided to look at more complete model that accounted for some of the dimerizations the proteins undergo:
Parameters
- a= production of NifA
- kf= binding rate of NifA and NifL
- kr= dissociation rate of NifA/NifL complex
- any d= degradation rate of that species
- kb:rate of complex formation (i.e. dimerization, formation of tetramers/hexamers)
- kb2: dissociation rate of such complexes given above
- m= represents how many subunits of NifA bind to NifHp (i.e., if we modeled NifA as a dimer [so 'i'=2], we would say m=3, since 2*3=6 (NifA is a hexamer).
- tmax: Maximal transcription rate
- tl: translation rate
Functions
- We model the production function of mRNA as a Hill-like, sigmoidal function
Variables
- mL=NifL mRNA
- L=NifL protein
- L2= NifL dimer
- A= NifA protein
- A2i= NifA complex, where i=1 is a dimer, i=4 is a tetramer, and i=6 is a hexamer
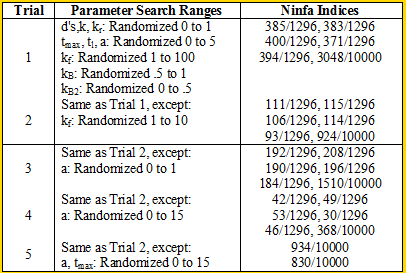
We ran some Ninfa Index simulations on the i=1 model (the other models were too complex to run the simulations on [my MacBook lacks computing power]):
Overall, we see smaller indices than the ones we saw in Model 1. Nonetheless,we see that the clock has at least some chance of oscillating.
When we decrease the randomization range of kf to in between 0 and 10 for trial 2, we see an almost threefold to fourfold decrease in the Ninfa index. Comparing trials 2 and 3 to Trial 1, we see that if we disturb the "equilibrium" of the system by producing lots of NifA (in trial 3) or too little Nifa (in trial 1), we can lose a lot of oscillatory potential. We seen increase in the Ninfa index when we "balance" the system by increasing range of the maximal transcription rate of NifL, as seen in Trial 4.
Back to Modeling
|
 "
"