Team:HKUSTers/Project
From 2008.igem.org
(→Reciprocal inhibiton) |
|||
| Line 158: | Line 158: | ||
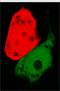
[[Image:greenred.jpg|100px|left|frameless]]The signal output is shown by green or red fluorescent protein (GFP/RFP) respectively. | [[Image:greenred.jpg|100px|left|frameless]]The signal output is shown by green or red fluorescent protein (GFP/RFP) respectively. | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 03:19, 30 October 2008
| Home | The Team | The Project | Parts to the Registry | Modelling | Notebook | Gallery |
The Design
"Making the dice"
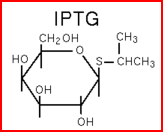
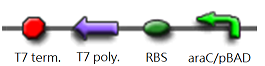
We "tell" our cells to start making the "dice"--T7 polymease by applying a pulse of IPTG. IPTG can activate the araC/pBAD promoter which leads to transcription of T7 promoter.
"Throwing the dice"
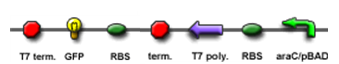
The core part of the randomizer consists of a pair of overlapping T7 promoters having similar binding affinities. The T7 polymerase binds randomly onto either the left or the right promoter to give an either 0 or 1 signal (reprenting by giving RFP or GFP).The binding is mutually exclusive, for if one binds to the promoter, it will block the binding of the other one as the two T7 promoters are overlapping to each other.
Reciprocal inhibiton
Then there are TetR and C1434 repressor repressor genes on both sides respectively. A repressor protein can be produced to interrupt transcription on the other side of the circuit.
For example, say, the T7 polymerase binds onto the promoter on the left side. The TetR repressor on the left, which was coded by TetR repressor gene, will bind to TetR operator on the right and repress transcription of right side of the promoter. Thus, the intial random event (binding of polymease) can be captured and amplified in a postive feedback circuit.
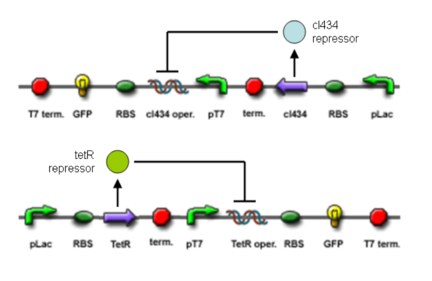
Final outcome
- Continous production of GFP if the polymease binds to the left of overlapped T7 promoter
- Continous production of RFP if the polymease binds to the right of overlapped T7 promoter
The signal output is shown by green or red fluorescent protein (GFP/RFP) respectively.
Construction details
Testing
Strain and vector used
- Strain: E. coli BBa_V1001 DH5a, with genotype
F-φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk- mk+ phoA supE44 λ- thi-1 gyrA96 relA1)
- Vector: pUC18+ λ-InCh2
Integration system
We have chosen the λInCh system as the vehicle for intergration.
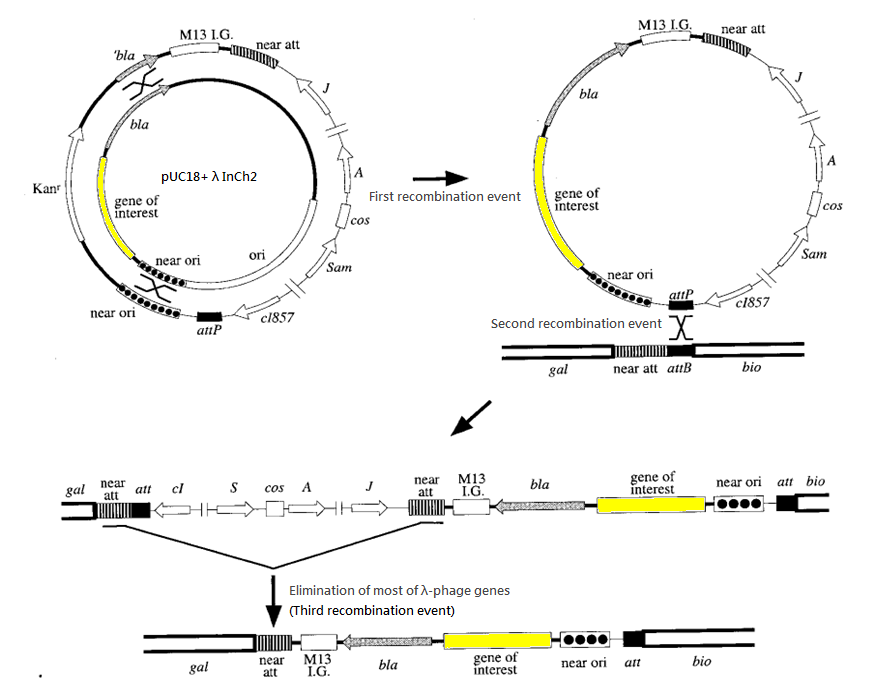
The success rate for recombination is about 1 in 1000 cells. Ampicillin is used to screen for the second recombination event and temperatute of 42 degree Celsius is used to screen for the third recombination event. (Such temperature deactivates the cl857 repressor and hence activates the harmful gene of λ-phage introduced in second recombination event, killing the host cells.)
The advantages of this approach include:
- No risk of >1 copy of target gene in our machine, so as to give a sharp output instead of "blurred" output caused by multiple copies
- Stable integration of a single copy of the construct DNA fragment
- Most ordinary E. coli strains and a variety of pBR322-derived Amp-resistant plasmids can be used
- λ-phage as the only specialized vector required
- Source:
Boyd D., Weiss D.S., Chen J.C., Beckwith J., Towards single-copy gene expression systems making gene cloning physiologically relevant: Lambda InCh, a simple Escherichia coli plasmid- chromosome shuttle system
(2000) Journal of Bacteriology, 182 (3), pp. 842-847. "
"