Team:Michigan/Project/Fabrication
From 2008.igem.org
(Difference between revisions)
| Line 63: | Line 63: | ||
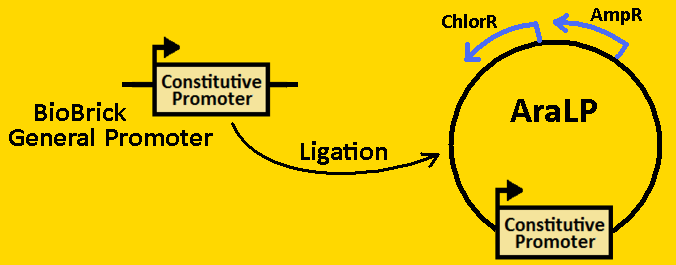
*Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | *Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | ||
*Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | *Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | ||
| - | *Analytical gel was then run to check fragment size of approximately 50bp. | + | *[https://2008.igem.org/Team:Michigan/Notebook/GelProtocol Analytical gel] was then run to check fragment size of approximately 50bp. |
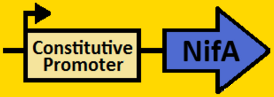
<br><br><font size=3 color=royalblue>NifA</font> | <br><br><font size=3 color=royalblue>NifA</font> | ||
| Line 76: | Line 76: | ||
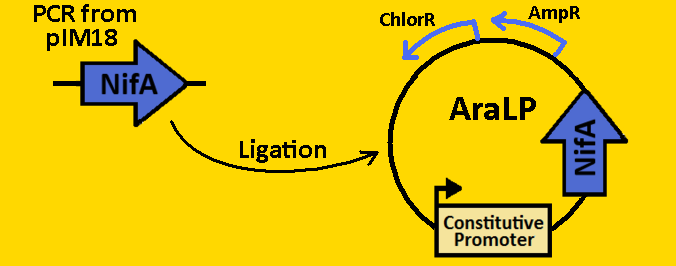
*Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | *Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | ||
*Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | *Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | ||
| - | *Analytical gel was then run to check fragment size of approximately 1650bp. | + | *[https://2008.igem.org/Team:Michigan/Notebook/GelProtocol Analytical gel] was then run to check fragment size of approximately 1650bp. |
| Line 102: | Line 102: | ||
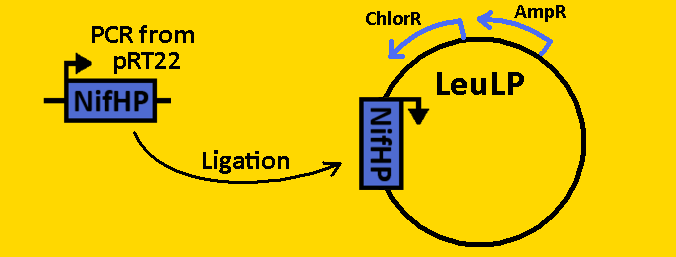
*Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | *Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | ||
*Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | *Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | ||
| - | *Analytical gel was then run to check fragment size of approximately 300bp. | + | *[https://2008.igem.org/Team:Michigan/Notebook/GelProtocol Analytical gel] was then run to check fragment size of approximately 300bp. |
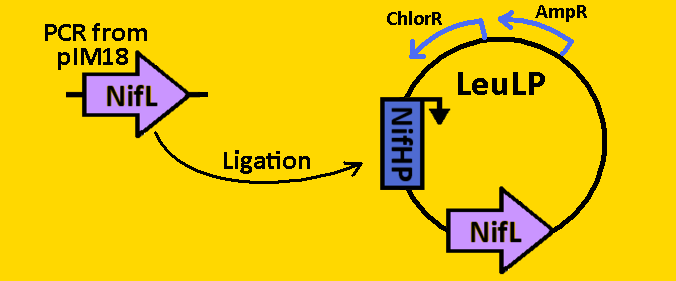
<br><br><font size=3 color=royalblue>NifL</font> | <br><br><font size=3 color=royalblue>NifL</font> | ||
| Line 115: | Line 115: | ||
*Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | *Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | ||
*Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | *Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | ||
| - | *Analytical gel was then run to check fragment size of approximately 2000bp. | + | *[https://2008.igem.org/Team:Michigan/Notebook/GelProtocol Analytical gel] was then run to check fragment size of approximately 2000bp. |
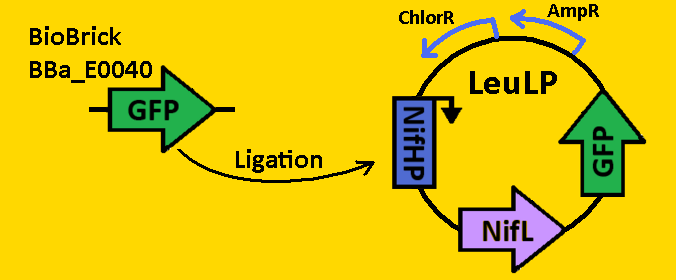
<br><br><font size=3 color=royalblue>GFP </font> | <br><br><font size=3 color=royalblue>GFP </font> | ||
| Line 126: | Line 126: | ||
*Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | *Plasmids from cells were [https://2008.igem.org/Team:Michigan/Notebook/PlasmidPrepProtocol isolated] by using QIAGEN MiniPrep Kit | ||
*Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | *Plasmids were then [https://2008.igem.org/Team:Michigan/Notebook/DigestProtocol digested] with EcoRI and SpeI overnight in 37⁰C (to ensure proper ligation). | ||
| - | *Analytical gel was then run to check fragment size of approximately 2700bp. | + | *[https://2008.igem.org/Team:Michigan/Notebook/GelProtocol Analytical gel] was then run to check fragment size of approximately 2700bp. |
!width="10%" align="justify" valign="top" style="background:transparent; color:navy"| | !width="10%" align="justify" valign="top" style="background:transparent; color:navy"| | ||
Revision as of 03:21, 30 October 2008
|
|---|
|
Project FabricationWe will be using landing pads to insert the Sequestillator onto the chromosome of E. coli. Noisy behavior has proven detrimental to clock studies throughout the past, and we hope to reduce the noise in our system using landing pads.
Landing Pad Plans for Sequestillator
Specific Fabrication TechniquesActivator Operon
Repressor Operon
|
|---|
 "
"