Team:NTU-Singapore/Modelling/Stochastic Modeling
From 2008.igem.org
Lalala8585 (Talk | contribs) |
Lalala8585 (Talk | contribs) |
||
| Line 18: | Line 18: | ||
=Stochastic Model For E7 + Imm Production= | =Stochastic Model For E7 + Imm Production= | ||
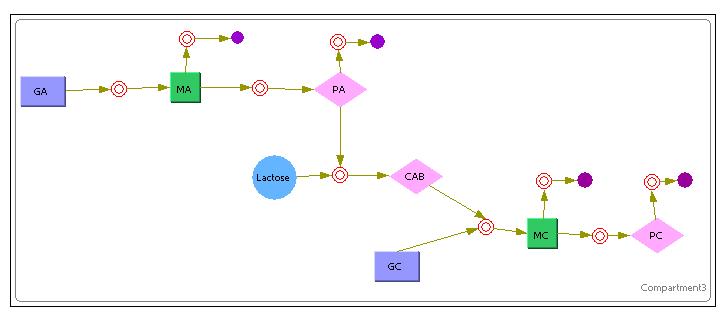
[[Image:Sys1.JPG|800px|System 1]]<br> | [[Image:Sys1.JPG|800px|System 1]]<br> | ||
| - | The above image shows the model that was used in Cellware for the 1st System. | + | The above image shows the model that was used in Cellware for the 1st System. <br> |
| - | + | The Purple Rectangles represent the DNA.<br> | |
| + | The Green Rectangles represent the mRNA.<br> | ||
| + | The Pink Diamonds represent the Protein.<br> | ||
| + | The Red Circles represent a certain reaction(Mass Action Reactions only since its a requirement for Stochastic simulations)<br> | ||
| + | The Purple Full Circles represented degradation of a particular substance.<br> | ||
Unlike the previous Simulink deterministic models, Cellware does not allow the user to inject certain variables at stipulated times. Therefore, the model shows inoculation of Lactose at the VERY START of the simulation. This is equivalent to exposing a newly divided cell to an environment of lactose rather than allowing it time to reach a certain steady state for its constitutive proteins. Nevertheless, we hope that the model can still allow us some insights into the system.<br> | Unlike the previous Simulink deterministic models, Cellware does not allow the user to inject certain variables at stipulated times. Therefore, the model shows inoculation of Lactose at the VERY START of the simulation. This is equivalent to exposing a newly divided cell to an environment of lactose rather than allowing it time to reach a certain steady state for its constitutive proteins. Nevertheless, we hope that the model can still allow us some insights into the system.<br> | ||
Revision as of 13:27, 12 July 2008
|
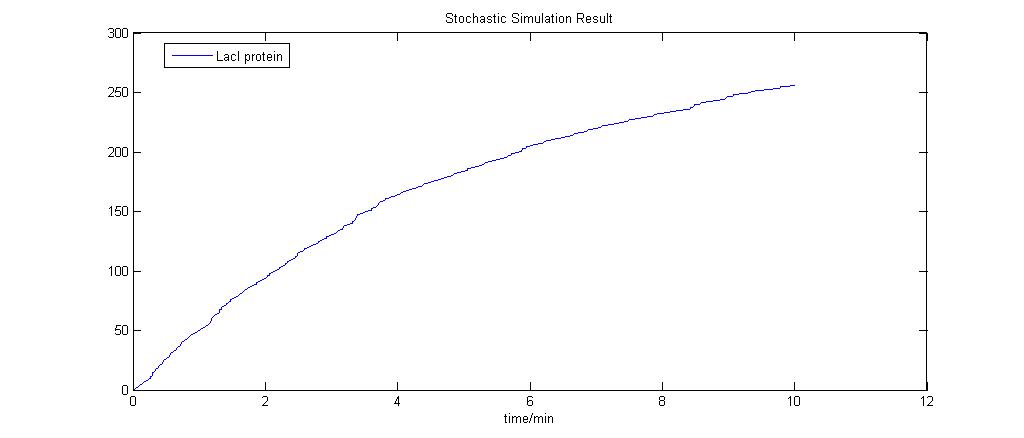
Preliminary Run
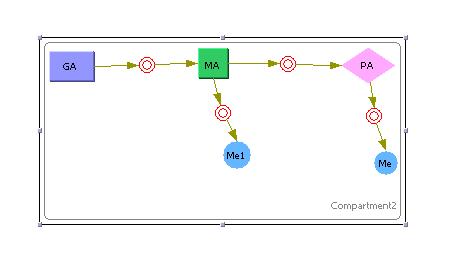
| 
|
This model was generated using the program [http://www.cellware.org/index.html CellWare]. The above model was simualted using a Hybrid ODE solver thats uses both deterministic and stochastic solvers. Work is now being carried out to see if this program can be used to build and simulate our systems.
All models below have been created using Cellware. [http://x.amath.unc.edu:16080/BioNetS/ Bionets2] is another option but we would first explore the use of the former program. The outputs are exported to MATLAB for plotting and visualisation.
Stochastic Model For E7 + Imm Production
The above image shows the model that was used in Cellware for the 1st System.
The Purple Rectangles represent the DNA.
The Green Rectangles represent the mRNA.
The Pink Diamonds represent the Protein.
The Red Circles represent a certain reaction(Mass Action Reactions only since its a requirement for Stochastic simulations)
The Purple Full Circles represented degradation of a particular substance.
Unlike the previous Simulink deterministic models, Cellware does not allow the user to inject certain variables at stipulated times. Therefore, the model shows inoculation of Lactose at the VERY START of the simulation. This is equivalent to exposing a newly divided cell to an environment of lactose rather than allowing it time to reach a certain steady state for its constitutive proteins. Nevertheless, we hope that the model can still allow us some insights into the system.
Before we actually use the program to run any form of stochastic simulations, it would be prudent to check how its deterministic models would turn out. By varying the amount of Lactose input, a few graphs were obtained and they are as shown.
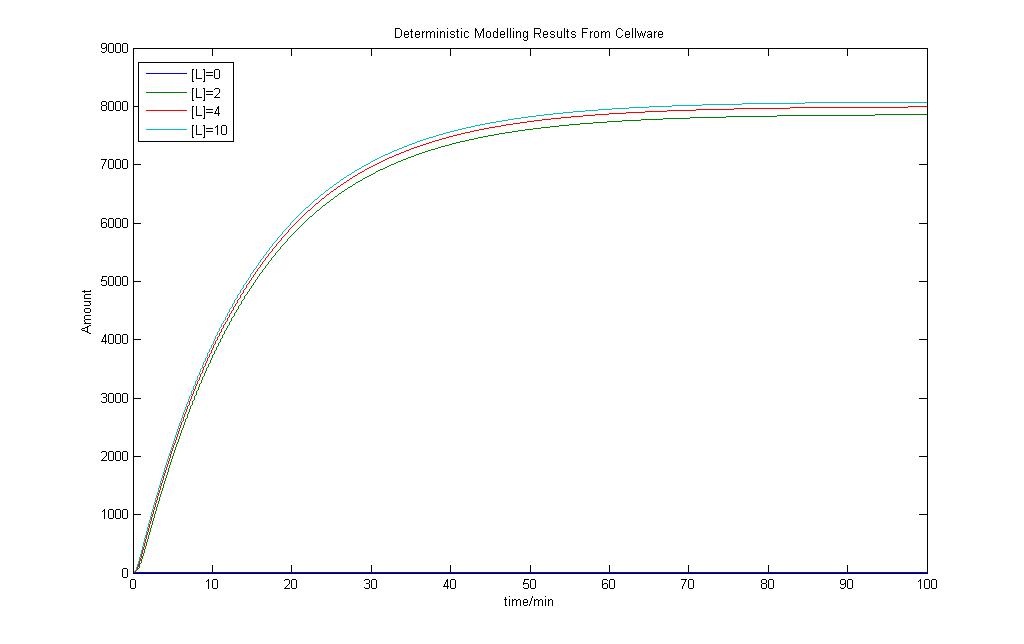
The model shows that there are differences in the output when the Lactose is changed but they are not exactly significant. However the model still shows very similar behaviour to the model we had obtained using Simulink earlier.
 "
"