Team:Groningen/Project
From 2008.igem.org
Aukevanheel (Talk | contribs) (→Overall project) |
Aukevanheel (Talk | contribs) (→Project Details) |
||
| Line 32: | Line 32: | ||
|[[Image:Grid.jpg|500px]] | |[[Image:Grid.jpg|500px]] | ||
| | | | ||
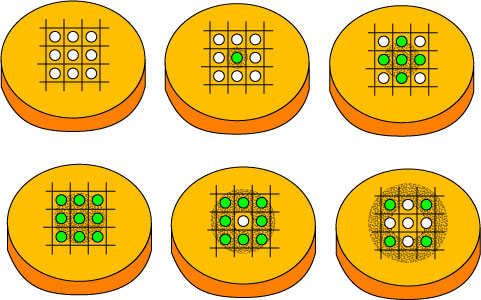
| - | + | This image depicts an example of an simple physical setup which could be used to "play" Conway's game of life. The green and white circles indicate colonies. HSL (the signal) can diffuse through the solid agar plates. The colonies should be able to act on the signal, and turn 'on' (green) or 'off' (white). | |
| | | | ||
|} | |} | ||
Revision as of 11:27, 16 September 2008
This year is the first year the University of Groningen is joining the iGEM competition. Our team – The Groningen Transformers – will focus on cell-cell signal systems, cellular automata and the Conway’s Game of Life!
Overall project
Since we have a multidisciplinary team we choose a project where lab work and modeling have to work together, for example by measuring parameters and optimizing our model every day. In our brainstorm sessions we all agreed that it would be fun to create a system in Escherichia coli which could repeat itself after one initial pulse. A cellular automata is a system in which rules are applied to cells and their neighbors in a two dimensional grid. We would like to create patterns and behaviors with simple rules. It is very important that complex behavior can be produced by simple rules. A very interesting example of a cellular automata is the Game of Life, invented by the mathematician John Conway in 1970. In this game grid cells can be live or dead. The most common rules are: with 2 or 3 neighbors a grid cell stays alive, with more or less it dies and with 3 neighbors a grid cell becomes alive. See figure x.
It will be a great challenge to make this game with a biological system. We already thought of a combination of bio bricks (link floor met parts voor ons systeem) and the necessary parts we have to add ourselves. Because this project is quite ambitious we will first try to make the subparts for the system.
The value of this game for biology is that we can synchronize cells better. Nowadays synchronization can be done by nitrogen starvation for example.
Project Details
In Conway's game of life cells have two states, in our case GFP expression or no GFP expression. Therefore we need a interval switch. We make this with a HSL/cI hybrid promoter that can be activated by a low HSL concentration; producing GFP and a promoter which will be switched on at higher HSL concentration; producing cI to block the first promoter, see figure x. This quorum sensing system was first discovered in Vibrio fischeri [Greenberg, 1999). The cells have to send HSL molecules to other cells. This double threshold switch makes the cells either ‘dead’; no GFP expression or ‘alive’; GFP expression and thereby forming our pattern on a two dimensional grid.
The devices we need to play the Conway's game of life in a cell or colony are:
- An interval device - Here cells need to know from its neighbors if they are 'dead' or 'alive', and when alive if its not overcrowded.
- A sender device - Cells need to let others known in what state they are: 'dead' or 'alive'.
- A receiver device - Cells need to obtain and process information from their neighbors.
- A reset function - After it gets overcrowded around one cell, it needs to reset, become 'dead'.
In the Registry of Standard Parts a lot of biobricks are involved in the [http://partsregistry.org/AHL AHL quorum sensing] system of Vibrio fischeri. For our system this was really helpful, since we did not want to use a cell-cell signal system which would interfere with E. coli's own quorum sensing system. We could use these bricks to create a system where cells will communicate with eachother and sense it at the same time. Moreover quite some other repressors and activators are present in the Registry. We chose to use the parts from the Registry as basic elements in the system. For some specific features we need to make and add them ourselves.
 "
"