Team:KULeuven/18 August 2008
From 2008.igem.org
(Difference between revisions)
(→Wet Lab) |
m |
||
| Line 1: | Line 1: | ||
| - | {{:Team:KULeuven/Tools/ | + | {{:Team:KULeuven/Tools/Header}} |
| - | + | ||
| - | + | ||
== Lab Work == | == Lab Work == | ||
| Line 25: | Line 23: | ||
== Remarks == | == Remarks == | ||
[[Image:TGS2.PNG | center]] | [[Image:TGS2.PNG | center]] | ||
| + | |||
| + | {{:Team:KULeuven/Tools/New_Day/Date_Retriever}} | ||
Revision as of 16:06, 18 September 2008
dock/undock dropdown
Contents |
Lab Work
Wet Lab
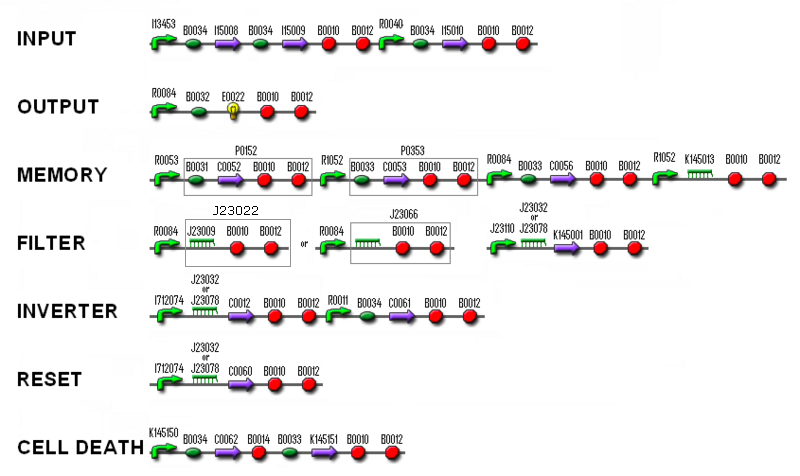
- We did some PCRs to prepare the following (new) BioBricks: K145014 (T7 polymerase with UmuD), K145013 (antisense LuxI) and K145151 (ccdB).
- We made a few digests: F1610 and B0015 were cut with EcoRI and XbaI, and J23100+B0032 and R0062+B0032 were cut with EcoRI and SpeI.
- We electroporated the following ligated parts: C0040+B0015, K145015+B0015, C0012+B0015 and C0060+B0015.
- We continued ligating: J23116+B0032, R0040+B0032, R0040+J23022, K145001+pSB1A2, K145001+B0015, R0084+B0032 and (R0084+J23022)+(J23109+J23032). this time, B0032 was first dephosphorylated.
Dry Lab
Modeling
Association and dissociation rates have been reinvestigated (HSL with LuxR), wrong values have been removed, LuxR autoactivation has been further incorporated; we've also included diffusion of HSL.
Wiki
Small updates for the wiki: adjusting pictograms, home page, ...
Started defining a global css, intermediate result here: Team:KULeuven/Zandbak/Global
Remarks
| << return to notebook | return to homepage >> | ||
| < previous friday | ← yesterday | tomorrow → | next monday > |
 "
"