Imperial College/24 September 2008
From 2008.igem.org
(Difference between revisions)
(New page: ==Dry Lab== ===Modelling Motility=== *Managed to run trajectory modelling on 90 cells yesterday. *Manual segmentation of runs done today to obtain a better curve fit. Hence certain cells...) |
(→Modelling Motility) |
||
| Line 6: | Line 6: | ||
*Manual segmentation of runs done today to obtain a better curve fit. Hence certain cells will have two or more runs, and two or more parameter sets respectively. | *Manual segmentation of runs done today to obtain a better curve fit. Hence certain cells will have two or more runs, and two or more parameter sets respectively. | ||
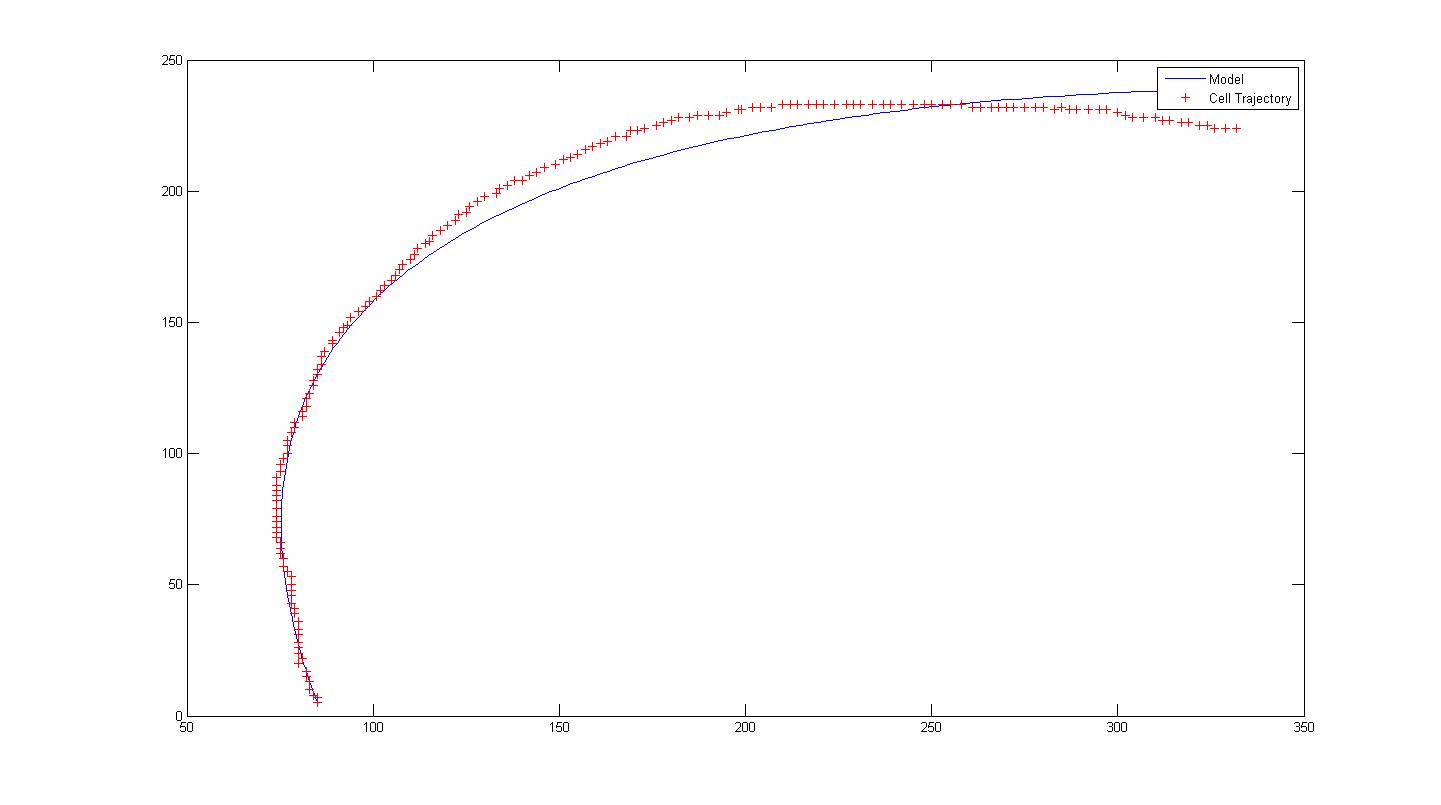
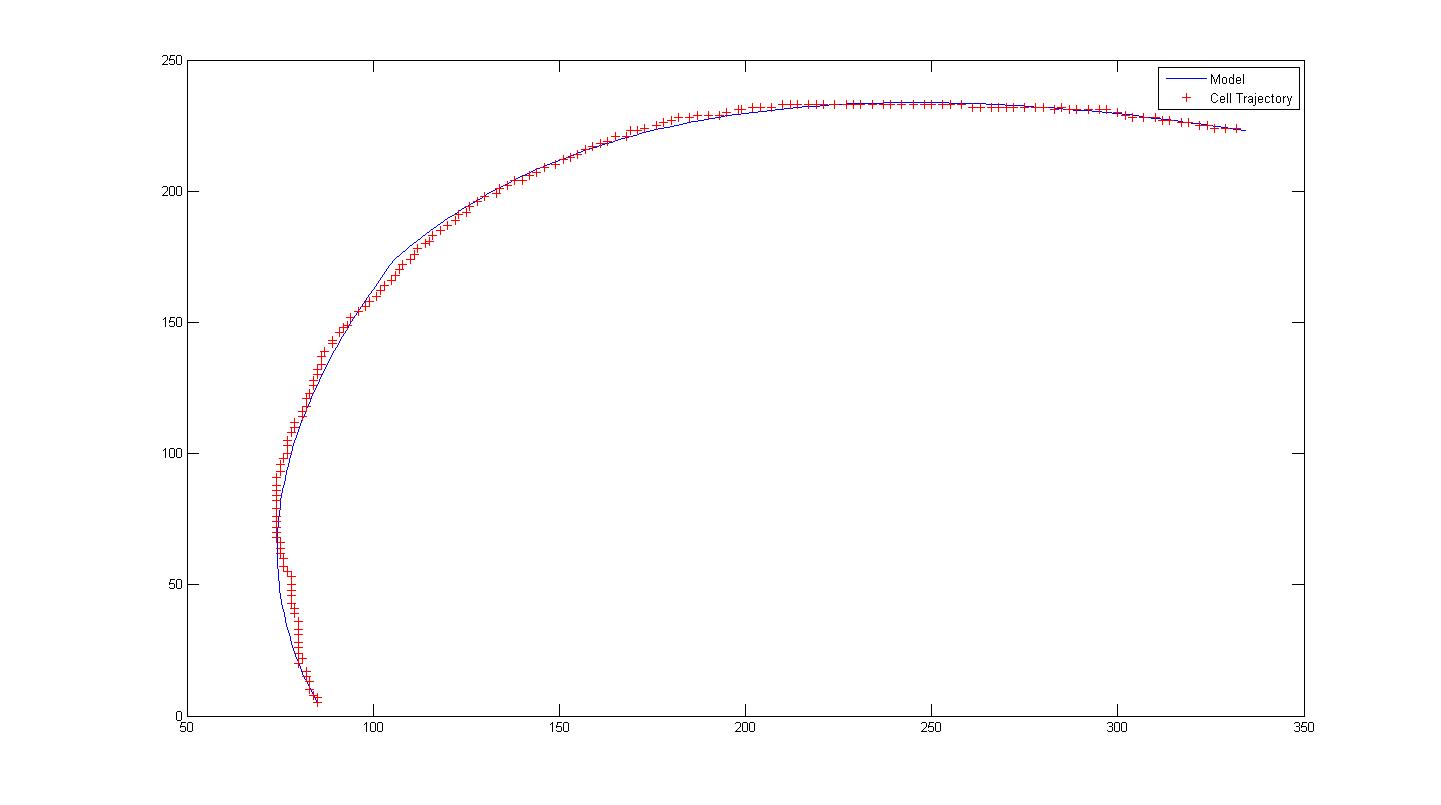
*Example is shown with 100908 Video 15 Cell 6. Notice that the model for a single run does not fit the data properly. Hence we introduce a change in orientation (parameters) at frame 75. This results in two separate runs, each with its respective set of parameters A, B and alpha. | *Example is shown with 100908 Video 15 Cell 6. Notice that the model for a single run does not fit the data properly. Hence we introduce a change in orientation (parameters) at frame 75. This results in two separate runs, each with its respective set of parameters A, B and alpha. | ||
| + | |||
| + | [[Image:V15Cell6.jpg|center|200px]] [[Image:V15Cell6(2).jpg|center|200px]] | ||
Revision as of 11:07, 24 September 2008
Dry Lab
Modelling Motility
- Managed to run trajectory modelling on 90 cells yesterday.
- Manual segmentation of runs done today to obtain a better curve fit. Hence certain cells will have two or more runs, and two or more parameter sets respectively.
- Example is shown with 100908 Video 15 Cell 6. Notice that the model for a single run does not fit the data properly. Hence we introduce a change in orientation (parameters) at frame 75. This results in two separate runs, each with its respective set of parameters A, B and alpha.
 "
"