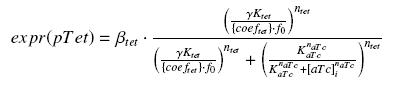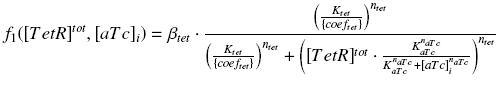Team:Paris/Modeling/f1
From 2008.igem.org
(Difference between revisions)
| Line 15: | Line 15: | ||
|unit | |unit | ||
|value | |value | ||
| + | |comments | ||
|- | |- | ||
|[expr(pTet)] | |[expr(pTet)] | ||
|expression rate of <br> pTet '''with RBS E0032''' | |expression rate of <br> pTet '''with RBS E0032''' | ||
|nM.min<sup>-1</sup> | |nM.min<sup>-1</sup> | ||
| + | | | ||
|see [[Team:Paris/Modeling/Programs|"findparam"]] <br> need for 20 measures | |see [[Team:Paris/Modeling/Programs|"findparam"]] <br> need for 20 measures | ||
|- | |- | ||
| Line 25: | Line 27: | ||
|min<sup>-1</sup> | |min<sup>-1</sup> | ||
|0.0198 | |0.0198 | ||
| + | |Time for Cell Division : 35 min. ; No degradation | ||
|- | |- | ||
|[GFP] | |[GFP] | ||
|GFP concentration at steady-state | |GFP concentration at steady-state | ||
|nM | |nM | ||
| + | | | ||
|need for 20 measures | |need for 20 measures | ||
|- | |- | ||
| Line 34: | Line 38: | ||
|value of the observed fluorescence | |value of the observed fluorescence | ||
|au | |au | ||
| + | | | ||
|need for 20 measures | |need for 20 measures | ||
|- | |- | ||
| Line 40: | Line 45: | ||
|nM.au<sup>-1</sup> | |nM.au<sup>-1</sup> | ||
|(1/79.429) | |(1/79.429) | ||
| + | | | ||
|} | |} | ||
| Line 49: | Line 55: | ||
|unit | |unit | ||
|value | |value | ||
| + | |comments | ||
|- | |- | ||
|β<sub>tet</sub> | |β<sub>tet</sub> | ||
|production rate of pTet '''with RBS E0032''' <br> β<sub>1</sub> | |production rate of pTet '''with RBS E0032''' <br> β<sub>1</sub> | ||
|nM.min<sup>-1</sup> | |nM.min<sup>-1</sup> | ||
| + | | | ||
| | | | ||
|- | |- | ||
| - | |( | + | |(K<sub>tet</sub>/{coef<sub>tetR</sub>}) |
|activation constant of pTet <br> K<sub>20</sub> | |activation constant of pTet <br> K<sub>20</sub> | ||
|nM | |nM | ||
| | | | ||
| + | |The [[Team:Paris/Modeling/Codes|optimisation program]] will give us (γ K<sub>tet</sub> / {coef<sub>tetR</sub>} ƒ0) | ||
| + | ||The litterature [[Team:Paris/Modeling/Bibliography|[?] ]] gives K<sub>tet</sub> = | ||
|- | |- | ||
|n<sub>tet</sub> | |n<sub>tet</sub> | ||
| Line 64: | Line 74: | ||
|no dimension | |no dimension | ||
| | | | ||
| + | |The litterature [[Team:Paris/Modeling/Bibliography|[?] ]] gives n<sub>tet</sub> = | ||
|- | |- | ||
|K<sub>aTc</sub> | |K<sub>aTc</sub> | ||
| Line 69: | Line 80: | ||
|nM | |nM | ||
| | | | ||
| + | |The litterature [[Team:Paris/Modeling/Bibliography|[?] ]] gives K<sub>aTc</sub> = | ||
|- | |- | ||
|n<sub>aTc</sub> | |n<sub>aTc</sub> | ||
| Line 74: | Line 86: | ||
|no dimension | |no dimension | ||
| | | | ||
| + | |The litterature [[Team:Paris/Modeling/Bibliography|[?] ]] gives n<sub>aTc</sub> = | ||
|} | |} | ||
Revision as of 13:13, 9 October 2008
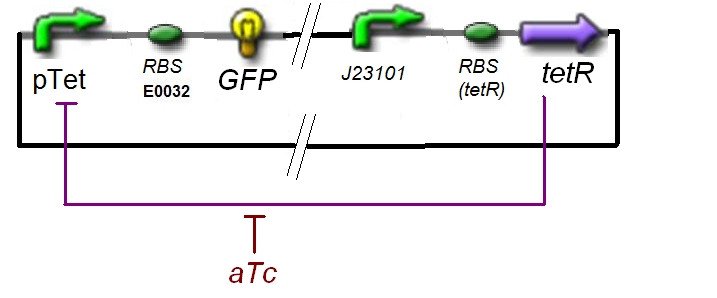
The experience would give us
Thus, at steady-state and in the exponential phase of growth :
| param | signification | unit | value | comments |
| [expr(pTet)] | expression rate of pTet with RBS E0032 | nM.min-1 | see "findparam" need for 20 measures | |
| γGFP | dilution-degradation rate of GFP(mut3b) | min-1 | 0.0198 | Time for Cell Division : 35 min. ; No degradation |
| [GFP] | GFP concentration at steady-state | nM | need for 20 measures | |
| (fluorescence) | value of the observed fluorescence | au | need for 20 measures | |
| conversion | conversion ration between fluorescence and concentration | nM.au-1 | (1/79.429) |
| param | signification corresponding parameters in the equations | unit | value | comments | |
| βtet | production rate of pTet with RBS E0032 β1 | nM.min-1 | |||
| (Ktet/{coeftetR}) | activation constant of pTet K20 | nM | The optimisation program will give us (γ Ktet / {coeftetR} ƒ0) | The litterature [?] gives Ktet = | |
| ntet | complexation order of pTet n20 | no dimension | The litterature [?] gives ntet = | ||
| KaTc | complexation constant aTc-TetR K19 | nM | The litterature [?] gives KaTc = | ||
| naTc | complexation order aTc-TetR n19 | no dimension | The litterature [?] gives naTc = |
Also, this experiment will allow us to know the expression of ƒ1 :
 "
"