User:Jec105
From 2008.igem.org
m |
m |
||
| Line 4: | Line 4: | ||
{{Imperial/Box2|Our Approach|[[Image:Cycle.PNG|center|500px]]|}} | {{Imperial/Box2|Our Approach|[[Image:Cycle.PNG|center|500px]]|}} | ||
| - | {{Imperial/Box1| | + | {{Imperial/Box1|Design| |
[[Image:Imperial_2008_Basic_Circuit.png|300px|right]] | [[Image:Imperial_2008_Basic_Circuit.png|300px|right]] | ||
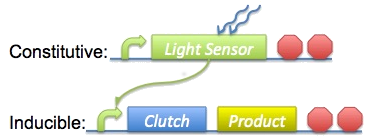
Our basic approach was to constitutively upregulate a light-sensitive pathway in ''B. subtilis'', then use a downstream product from that pathway to selectively regulate expression of our clutch and biomaterial. This overview is shown on the right. To do this we need to produce at least one constitutive promoter to precede our light-sensing mechanism, as well as a 'light-inducible' promoter in front of our clutch and biomaterial. Additionally, we'll need to make all the other standard parts such as ribosome binding sites (RBS) and terminators. | Our basic approach was to constitutively upregulate a light-sensitive pathway in ''B. subtilis'', then use a downstream product from that pathway to selectively regulate expression of our clutch and biomaterial. This overview is shown on the right. To do this we need to produce at least one constitutive promoter to precede our light-sensing mechanism, as well as a 'light-inducible' promoter in front of our clutch and biomaterial. Additionally, we'll need to make all the other standard parts such as ribosome binding sites (RBS) and terminators. | ||
Revision as of 21:35, 20 October 2008
Summer Summary
|
|||||||||||||||||||||||||||
 "
"