Team:Caltech/Project
From 2008.igem.org
| Line 52: | Line 52: | ||
{{!}} | {{!}} | ||
As more complicated and interconnected biological circuits are synthesized, there becomes the need for the simple integration of multiple functions into a single bacterial cell line. However, some of these functions may be incompatible or may kill the cell, such that each cell can only express a single function at any time and must be regenerated if the functionality is fatal. We aim to combine multiple mutually exclusive and potentially fatal functions into a single bacterial cell line that, as a population, expresses the entire set of functions. | As more complicated and interconnected biological circuits are synthesized, there becomes the need for the simple integration of multiple functions into a single bacterial cell line. However, some of these functions may be incompatible or may kill the cell, such that each cell can only express a single function at any time and must be regenerated if the functionality is fatal. We aim to combine multiple mutually exclusive and potentially fatal functions into a single bacterial cell line that, as a population, expresses the entire set of functions. | ||
| + | |||
In particular, we want only one of the four subprojects described above to be turned on in any given cell, or else the cell may be overburdened by our constructs. At the same time, we want our bacterial population to have all four abilities. In addition, each of the four subprojects causes the death of the host cell through self-induced lysis. We need a system that is able to combine all subprojects into one coherent system. | In particular, we want only one of the four subprojects described above to be turned on in any given cell, or else the cell may be overburdened by our constructs. At the same time, we want our bacterial population to have all four abilities. In addition, each of the four subprojects causes the death of the host cell through self-induced lysis. We need a system that is able to combine all subprojects into one coherent system. | ||
| + | |||
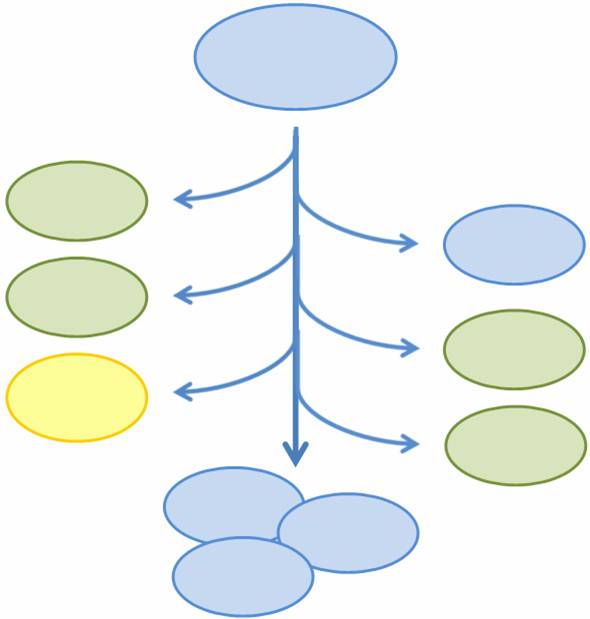
We propose a system in which bacterial cells initially start in an undifferentiated state and randomly differentiate into one of three possible final states. To build this system, we designed two genetic devices. One relies on DNA polymerase slippage upon replication of a long stretch of short nucleotide repeats, a phenomenon termed slipped-strand mutation (SSM). The other device uses the recombinase protein FimE to flip DNA segments. The two devices can form a system that permits the novel introduction of random multi-state differentiation into bacterial cells. Here, we present evidence that short nucleotide repeats can be used as a stochastic switch and that FimE activity depends on the length of the segment being flipped. | We propose a system in which bacterial cells initially start in an undifferentiated state and randomly differentiate into one of three possible final states. To build this system, we designed two genetic devices. One relies on DNA polymerase slippage upon replication of a long stretch of short nucleotide repeats, a phenomenon termed slipped-strand mutation (SSM). The other device uses the recombinase protein FimE to flip DNA segments. The two devices can form a system that permits the novel introduction of random multi-state differentiation into bacterial cells. Here, we present evidence that short nucleotide repeats can be used as a stochastic switch and that FimE activity depends on the length of the segment being flipped. | ||
{{!}}} | {{!}}} | ||
}} | }} | ||
Revision as of 23:02, 20 October 2008
|
People
|
Subprojects
Note: Click on the subproject title or picture for a detailed description of the subproject
Oxidative Burst
Phage Pathogen Defense
Lactose Intolerance
Vitamin Production
Population Variation
|
 "
"