Team:Caltech/Project/Lactose intolerance
From 2008.igem.org
RobertOvadia (Talk | contribs) |
RobertOvadia (Talk | contribs) |
||
| Line 42: | Line 42: | ||
==Parts== | ==Parts== | ||
| - | LacY in pSB1A2 BBa_K137001 | + | LacY in pSB1A2 BBa_K137001<br> |
| - | Lysis in pSB1A2 BBa_K137126 | + | Lysis in pSB1A2 BBa_K137126<br> |
| - | B0031 + LacY in pSB1A2 BBa_S03970 | + | B0031 + LacY in pSB1A2 BBa_S03970<br> |
| - | B0033 + LacY in pSB1A2 BBa_S03971 | + | B0033 + LacY in pSB1A2 BBa_S03971<br> |
| - | B0034-LacZ + B0015 in pSB1AK3 BBa_S03973 | + | B0034-LacZ + B0015 in pSB1AK3 BBa_S03973<br> |
| - | B0031-LacY + B0015 in pSB1AK3 BBa_S04107 | + | B0031-LacY + B0015 in pSB1AK3 BBa_S04107<br> |
| - | B0033-LacY + B0015 in pSB1AK3 BBa_S04108 | + | B0033-LacY + B0015 in pSB1AK3 BBa_S04108<br> |
| - | J23113 + B0031-LacY-B0015 in J61002 BBa_S04109 | + | J23113 + B0031-LacY-B0015 in J61002 BBa_S04109<br> |
| - | J23100 + B0031-LacY-B0015 in J61002 BBa_S04123 | + | J23100 + B0031-LacY-B0015 in J61002 BBa_S04123<br> |
| - | J23106 + B0031-LacY-B0015 in J61002 BBa_S04122 | + | J23106 + B0031-LacY-B0015 in J61002 BBa_S04122<br> |
| - | J23113 + B0033-LacY-B0015 in J61002 BBa_S04110 | + | J23113 + B0033-LacY-B0015 in J61002 BBa_S04110<br> |
| - | J23106 + B0033-LacY-B0015 in J61002 BBa_S04111 | + | J23106 + B0033-LacY-B0015 in J61002 BBa_S04111<br> |
| - | J23100 + B0033-LacY-B0015 in J61002 BBa_S04112 | + | J23100 + B0033-LacY-B0015 in J61002 BBa_S04112<br> |
| - | B0033-LacY SDM +H in pSB1A2 BBa_S04041 | + | B0033-LacY SDM +H in pSB1A2 BBa_S04041<br> |
| - | B0033-LacY SDM +2H in pSB1A2 BBa_S04022 | + | B0033-LacY SDM +2H in pSB1A2 BBa_S04022<br> |
| - | B0033-LacY SDM +H + B0034-LacZ-B0015 in pSB1AK3 BBa_S04113 | + | B0033-LacY SDM +H + B0034-LacZ-B0015 in pSB1AK3 BBa_S04113<br> |
| - | B0033-LacY SDM +2H + B0034-LacZ-B0015 in pSB1AK3 BBa_S04054 | + | B0033-LacY SDM +2H + B0034-LacZ-B0015 in pSB1AK3 BBa_S04054<br> |
| - | J23100 B0033-LacY SDM +2H + B0034-LacZ-B0015 in J61002 BBa_S04055 | + | J23100 B0033-LacY SDM +2H + B0034-LacZ-B0015 in J61002 BBa_S04055<br> |
| - | A81 (Lac. Po.) BBa_K137127 | + | A81 (Lac. Po.) BBa_K137127<br> |
| - | B4 (Lac. Po.) BBa_K137128 | + | B4 (Lac. Po.) BBa_K137128<br> |
| - | B0015 + A81 in pSB2K3 BBa_S04115 | + | B0015 + A81 in pSB2K3 BBa_S04115<br> |
| - | B0015 + B4 in pSB2K3 BBa_S04116 | + | B0015 + B4 in pSB2K3 BBa_S04116<br> |
| - | B0015 + A81 + Lysis + B0015 in pSB2K3 BBa_S04117 | + | B0015 + A81 + Lysis + B0015 in pSB2K3 BBa_S04117<br> |
| - | B0015 + B4 + Lysis + B0015 in pSB2K3 BBa_S04118 | + | B0015 + B4 + Lysis + B0015 in pSB2K3 BBa_S04118<br> |
| - | Lysis + B0015 in pSB2K3 BBa_S04114 | + | Lysis + B0015 in pSB2K3 BBa_S04114<br> |
Revision as of 20:27, 21 October 2008
|
People
|
Curing Lactose Intolerance
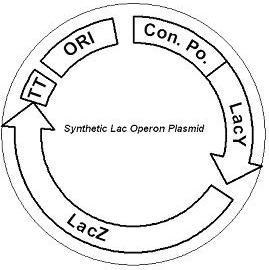
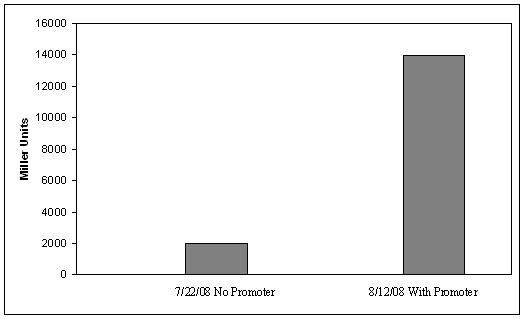
IntroductionThe gut flora of our digestive tract contains microorganisms that perform various useful functions for their hosts. Examples of such functions include growth inhibition of harmful microorganisms1, defense against the causes of many forms of Inflammatory Bowel Disease2, and the fermentation of carbohydrates and other molecules the human body cannot normally digest. Some bacterial strains, including the E. coli strain ‘Nissle 1917’, can persist in the gut of mice for months. Engineering these bacteria provide a new platform to treat various human diseases. System DesignLactose RegulatorThe first plasmid consists of a synthetic lactose operon under strong constitutive expression (Fig. 1). Our synthetic lactose operon encodes the ß-galactosidase LacZ and a lactose permease LacY. LacY is a membrane protein that actively transports lactose into the cell. Since most membrane proteins are toxic when overexpressed, we optimized our system to express appropriate levels of LacY without killing the cell. In the end, we want to express as much ß-galactosidase as we can to cleave as much lactose present in the large intestine. Lactose-Induced Lysis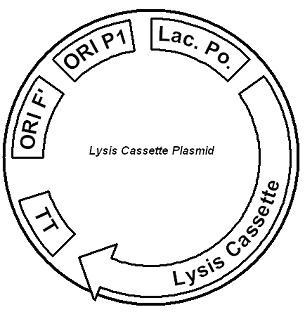 Figure 2. The lysis cassette plasmid acts as a lactose sensor. Intracellular lactose accumulation induces overexpression of the lysis cassette. Lactose inhibits binding of the LacI repressor to the promoter and P1 high copy origin of replication. A mutated holin gene in the lysis cassette allows faster lysis times. The second plasmid contains lactose inducible expression of λ phage lysis genes under a lactose inducible promoter (Fig. 2). To release ß-galactosidase, the cells will lyse when enough lactose is present in the cell to induce the expression of the lysis genes. It is important that the lactose-inducible promoter is tightly regulated since leaky expression will cause spontaneous lysis. To accomplish tight expression, specific lac promoters will keep our system from lysing5. In addition, the plasmid copy number remains low copy until induced with lactose, when the plasmid copy number increases to high copy. ResultsLactose Regulator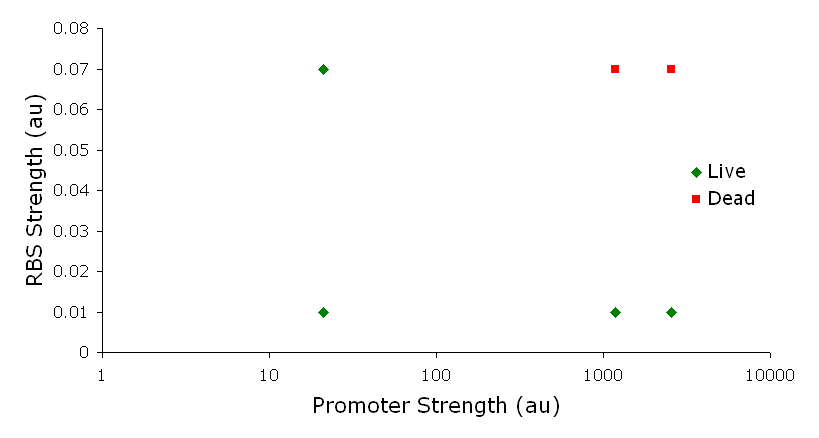 Figure 3. Varying the expression levels of LacY with different promoters and ribosome binding sites. Cells died at our two highest expression levels, but survived on the other four. The construct with the weakest RBS was selected since we could express LacZ in the same operon with a strong promoter. This combination allows our cell to express high levels of LacZ and non-toxic levels of LacY. In the first plasmid, it was discovered that LacY is toxic when overexpressed. To determine safe expression levels, constructs were built with varying levels of LacY expression. Six plasmids were built from combinations of three different promoters and two different ribosome binding sites (Fig. 3). The cells appeared to have died when expressing the strongest and medium strength promoters along with the strongest ribosome binding site. Our information was based off transformants, and the plates with no transformants were classified as dead cells due to high LacY expression. Based on these results, we decided to express our synthetic lac operon by combining the strongest constitutive promoter with the weakest RBS for LacY and the strongest RBS for LacZ, preventing LacY toxicity while expressing LacZ in large quantities. The ß-galactosidase assay was performed on the synthetic network containing the lactose operon (Fig. 4). The first assay was performed on a strain containing the plasmid lacking the strong constitutive promoter. This assay was performed to simulate the eventual system regulation. We saw significant levels of ß-galactosidase, a surprising result since the plasmid lacked a promoter. These levels are likely due to spurious transcription amplified by our strong ribosome binding site and high copy plasmid. We then repeated the assay with the strong constitutive promoter to observe the dynamic range of LacZ expression in this system.
Lactose-Induced LysisOur second plasmid was not finished, but necessary parts have been cloned for future construction. There remains one more cloning step to place the lysis cassette into the vector containing the promoters. The single base mutations in the lysis cassette were not made, and if time allowed, they would have been completed as the final step. DiscussionOur final system was not completely finished; however, future construction will be minimal. The synthetic lac operon plasmid was completely constructed with the insertion of two histidyl residues in their appropriate locations. Our lactose-induced lysis plasmid was one cloning step away from being completed. The promoter and flanking terminator were cloned in parallel with the lysis cassette and a terminator. The last cloning step would be to ligate the two together. In addition, the two separate point mutations have not been made, and the final step in our cloning would be to add those mutations.
MethodsMethods can be found here PartsLacY in pSB1A2 BBa_K137001
References
|
 "
"