Home
People
Project Details
Protocols
Completed Systems
Biosafety
Support
|
Curing Lactose Intolerance
Introduction
The gut flora of our digestive tract contains microorganisms that perform various useful functions for their hosts. Examples of such functions include growth inhibition of harmful microorganisms1, defense against the causes of many forms of Inflammatory Bowel Disease2, and the fermentation of carbohydrates and other molecules the human body cannot normally digest. Some bacterial strains, including the E. coli strain ‘Nissle 1917’, can persist in the gut of mice for months. Engineering these bacteria provide a new platform to treat various human diseases.
Lactose intolerance is characterized by the inability to break down lactose in the small intestine. The undigested lactose instead passes to the large intestine, leading to two negative processes: osmotic imbalance and bacterial fermentation. High lactose levels raise the osmolarity of the colon, causing diarrhea. In addition, gut microbes metabolize the lactose into methane gas, causing abdominal pain. Both problems must be addressed in order to fully treat lactose intolerance. If we simply have our strain metabolize lactose, another strain will further ferment the byproducts, resulting in the same side effects.
Instead, we have our engineered two plasmids which allow the host to uptake lactose, clearing the sugar from the colon. To do this, we have engineered the ‘Nissle 1917’ strain to release ß-galactosidase, an enzyme that cleaves lactose into glucose and galactose. Since protein secretion is difficult in E. coli, our cells were engineered to lyse in order to release ß-galactosidase. However, lysis must occur only when lactose is present in the gut. If lysis were to occur when lactose was absent, this would kill all our engineered cells at any given moment. The cells also express a lactose permease, allowing the strain to sense lactose in all conditions.
Figure showing before lysis, during lysis, and after lysis will be added soon.
System Design
Lactose Regulator
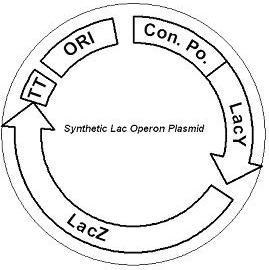 Figure 1. A mutated LacY allows the uptake of lactose while LacZ produces large amounts of ß-galactosidase. The plasmid contains a strong constitutive promoter regulating both genes and a ColE1 replication origin. The first plasmid consists of a synthetic lactose operon under strong constitutive expression (Fig. 1). Our synthetic lactose operon encodes the ß-galactosidase LacZ and a lactose permease LacY. LacY is a membrane protein that actively transports lactose into the cell. Since most membrane proteins are toxic when overexpressed, we optimized our system to express appropriate levels of LacY without killing the cell. In the end, we want to express as much ß-galactosidase as we can to cleave as much lactose present in the large intestine.
The human gut is an unpredictable environment, and we wish our engineered cells to behave reliably despite this variability. E. coli are unable to uptake lactose in the presence of glucose, a phenomenon known as carbon catabolite repression. Catabolite repression is mediated by Enzyme IIA Glucose (IIAGluc), which inhibits the uptake of lactose in the presence of glucose by binding to LacY3. Previous research has identified various LacY mutations that prevent this inhibition and achieve increased uptake of lactose in the presence of glucose4.
Lactose-Induced Lysis
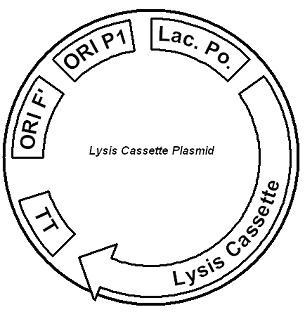 Figure 2. The lysis cassette plasmid acts as a lactose sensor. Intracellular lactose accumulation induces overexpression of the lysis cassette. Lactose inhibits binding of the LacI repressor to the promoter and P1 high copy origin of replication. A mutated holin gene in the lysis cassette allows faster lysis times. The second plasmid contains lactose inducible expression of λ phage lysis genes under a lactose inducible promoter (Fig. 2). To release ß-galactosidase, the cells will lyse when enough lactose is present in the cell to induce the expression of the lysis genes. It is important that the lactose-inducible promoter is tightly regulated since leaky expression will cause spontaneous lysis. To accomplish tight expression, specific lac promoters will keep our system from lysing5. In addition, the plasmid copy number remains low copy until induced with lactose, when the plasmid copy number increases to high copy.
Using wild type λ phage lysis genes, lysis occurs 40-45 minutes after induction by lactose. Decreasing the lag time will reduce the extent of lactose fermentation and therefore produce fewer deleterious effects. Previous research has uncovered mutations that shorten the lysis time to approximately 10-15 minutes6, and these mutations will be incorporated into our final construct.
Results
Lactose Regulator
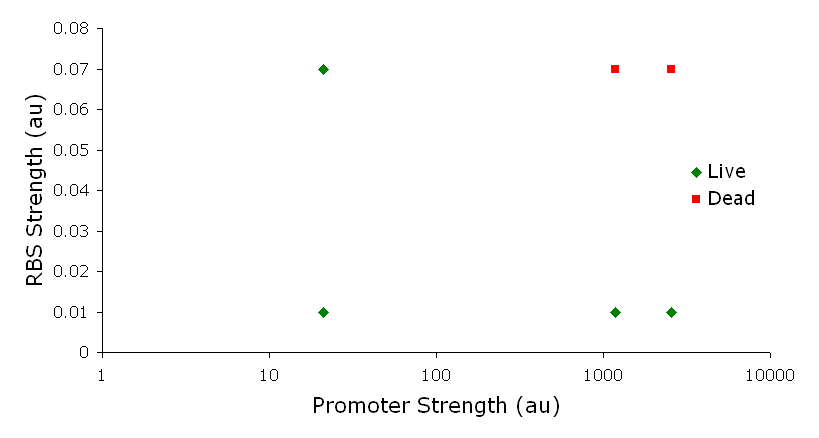 Figure 3. Varying the expression levels of LacY with different promoters and ribosome binding sites. Cells died at our two highest expression levels, but survived on the other four. The construct with the weakest RBS was selected since we could express LacZ in the same operon with a strong promoter. This combination allows our cell to express high levels of LacZ and non-toxic levels of LacY. In the first plasmid, it was discovered that LacY is toxic when overexpressed. To determine safe expression levels, constructs were built with varying levels of LacY expression. Six plasmids were built from combinations of three different promoters and two different ribosome binding sites (Fig. 3). The cells appeared to have died when expressing the strongest and medium strength promoters along with the strongest ribosome binding site. Our information was based off transformants, and the plates with no transformants were classified as dead cells due to high LacY expression. Based on these results, we decided to express our synthetic lac operon by combining the strongest constitutive promoter with the weakest RBS for LacY and the strongest RBS for LacZ, preventing LacY toxicity while expressing LacZ in large quantities.
As mentioned earlier, our cells had to inhibit carbon catabolite repression to ensure uptake of lactose under any condition. Various mutations, including the insertion of two histidyl residues between amino acids S194 and A195 (ref 4) in the LacY gene, prevent the inhibition by IIAGluc. These insertions have been made and will be tested in future studies.
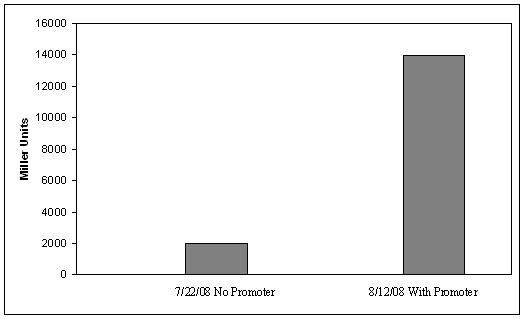 Figure 4. ß-galactosidase activity of plasmid #1, with and without the strong constitutive promoter. Error bars on the data with a promoter show the standard deviation of two measurements. Lactose-Induced Lysis
Our second plasmid was not finished, but necessary parts have been cloned for future construction. There remains one more cloning step to place the lysis cassette into the vector containing the promoters. The single base mutations in the lysis cassette were not made, and if time allowed, they would have been completed as the final step.
Discussion
Our final system was not completely finished; however, future construction will be minimal. The synthetic lac operon plasmid was completely constructed with the insertion of two histidyl residues in their appropriate locations. Our lactose-induced lysis plasmid was one cloning step away from being completed. The promoter and flanking terminator were cloned in parallel with the lysis cassette and a terminator. The last cloning step would be to ligate the two together. In addition, the two separate point mutations have not been made, and the final step in our cloning would be to add those mutations.
After completing the construction phase, we would characterize our system. Characterization will be completed on a construct containing our lactose inducible promoters and GFP. This would show our promoters can maintain tight expression of GFP from uninduced to induced. We would combine this construct with our first plasmid containing the mutated LacY, to show we can achieve induction by lactose even in the presence of glucose. To show the effect our lysis plasmid, it would be necessary to achieve lysis with lactose. In addition, we want to show that we can achieve lysis with lactose in the presence of glucose. Finally, the final assay we want to perform on our system would show our cells able to uptake lactose in the presence of glucose, and lyse soon after. Once the cells lyse, the cell lysis should contain ß-galactosidase at levels close to what we received from our ß-galactosidase assay on plasmid 1. Once characterization is completed, our construct would be moved to the ‘Nissle 1917’ strain where further characterization and modifications would take place.
Methods
Methods can be found here
Parts
| Registry Number | Plasmid | Part | Status
|
| [http://partsregistry.org/Part:BBa_K137002 BBa_K137002]
| pSB1A2 | LacY | Constructed
|
| [http://partsregistry.org/Part:BBa_K137126 BBa_K137126]
| pSB1A2 | Lysis Cassette | Constructed
|
| [http://partsregistry.org/Part:BBa_S03970 BBa_S03970]
| pSB1A2 | B0031 + LacY | Constructed
|
| [http://partsregistry.org/Part:BBa_S03971 BBa_S03971]
| pSB1A2 | B0033 + LacY | Constructed
|
| [http://partsregistry.org/Part:BBa_S03973 BBa_S03973]
| pSB1AK3 | B0034-LacZ + B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04107 BBa_S04107]
| pSB1AK3 | B0031-LacY + B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04108 BBa_S04108]
| pSB1AK3 | B0033-LacY + B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04109 BBa_S04109]
| J61002 | J23113 + B0031-LacY-B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04122 BBa_S04122]
| J61002 | J23106 + B0031-LacY-B0015 | Unavailable
|
| [http://partsregistry.org/Part:BBa_S04123 BBa_S04123]
| J61002 | J23100 + B0031-LacY-B0015 | Unavailable
|
| [http://partsregistry.org/Part:BBa_S04110 BBa_S04110]
| J61002 | J23113 + B0033-LacY-B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04111 BBa_S04111]
| J61002 | J23106 + B0033-LacY-B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04112 BBa_S04112]
| J61002 | J23100 + B0033-LacY-B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04041 BBa_S04041]
| pSB1A2 | B0033 + LacY +H | Constructed
|
| [http://partsregistry.org/Part:BBa_S04022 BBa_S04022]
| pSB1A2 | B0033 + LacY +2H | Constructed
|
| [http://partsregistry.org/Part:BBa_S04113 BBa_S04113]
| pSB1AK3 | B0033-LacY +H + B0034-LacZ-B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04054 BBa_S04054]
| pSB1AK3 | B0033-LacY +2H + B0034-LacZ-B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04055 BBa_S04055]
| J61002 | J23100 + B0033-LacY SDM +2H-B0034-LacZ-B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_K137124 BBa_K137124]
| pSB1A2 | LacI Repressed Promoter A81 | Constructed
|
| [http://partsregistry.org/Part:BBa_K137125 BBa_K137125]
| pSB1A2 | LacI Repressed Promoter B4 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04114 BBa_S04114]
| pSB2K3 | Lysis + B0015 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04115 BBa_S04115]
| pSB2K3 | B0015 + A81 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04116 BBa_S04116]
| pSB2K3 | B0015 + B4 | Constructed
|
| [http://partsregistry.org/Part:BBa_S04117 BBa_S04117]
| pSB2K3 | B0015-A81 + Lysis-B0015 | Constructed?
|
| [http://partsregistry.org/Part:BBa_S04118 BBa_S04118]
| pSB2K3 | B0015-B4 + Lysis-B0015 | Constructed?
|
References
- Guarner F and Malagelada JR. 2003. Gut flora in health and disease. The Lancet, Volume 361, Issue 9356, 8 February 2003, Pages 512-519.
- Sears CL. 2005. A dynamic partnership: Celebrating our gut flora. Anaerobe, Volume 11, Issue 5, Pages 247-251.
- Deutscher, J., Francke, C. and Postma, P.W. 2006. How Phosphotransferase System-Related Protein Phosphorylation Regulates Carbohydrate Metabolism in Bactertia. Microbial and Molecular Biology Reviews. 2006: 939-1031
- Hoischen, C., Levin, J., Pitaknarongphorn, S., Reizer, J., and Saier , M. H. Jr. 1996. Involvement of the central loop of the lactose permease of Escherichia coli in its allosteris regulation by the glucose-specific enzyme IIA of the phosphoenolpyruvate-dependent phosphotransferase system. J. Bacteriol. 178: 6082-6086
- Cox RS III, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 2007;3:145.
- Gründling, A., M. D. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. USA 98:9348-9352.
  |


 "
"


