Team:Chiba/Project
From 2008.igem.org
(→Signal Molecule Receiver) |
(→Quorum-Sensing Cross-talk) |
||
| Line 105: | Line 105: | ||
====Quorum-Sensing Cross-talk==== | ====Quorum-Sensing Cross-talk==== | ||
| - | [[Image: | + | [[Image:Receiver switch chiba.jpg|left]] |
English: | English: | ||
日本語: | 日本語: | ||
| - | + | [[Image:Table R chiba.gif|frame|left|Table.1 LuxR family gene Data modified from Fekete et.al. Anal Bioanal Chem (2007)]]<br clear="all"> | |
====Copy nomber of Receiver==== | ====Copy nomber of Receiver==== | ||
Revision as of 07:37, 26 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
Contents |
Abstract
- "Team : Chiba - E.coli time manager"
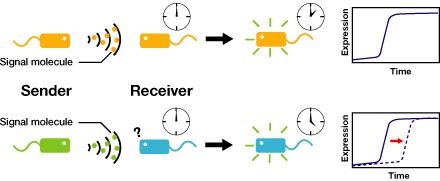
We control the timing of gene expression by using multiple signaling devices.To this end,we utilize molecules associated with Quorum sensing, a phenomenon that allows bacteria to communicate with each other.Our project uses two classes of bacteria: senders and receivers. Senders produce signaling molecules, and Receivers are activated only after a particular concentration of this molecule is reached.Although different quorum sensing species have slightly different signaling molecules, these molecules are not completely specific to their hosts and cross-species reactivity is observed [http://www3.interscience.wiley.com/journal/119124142/abstract (1)],[http://partsregistry.org/Part:BBa_F2620:Specificity (2)]. Communication using non-endogenous molecules is less sensitive, and requires a higher signal concentration to take effect.This results in slower activation of receivers.
Introduction
Our project
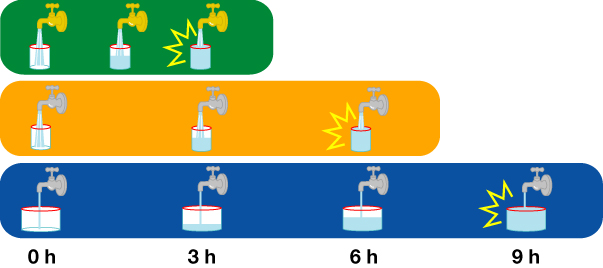
従来:消えていく物質の存在時間によって時間調節する。
千葉:常に物質が一定の割合で蓄積されるとき、あるスレッショルド超えるまでの時間を調節する。
調節する方法には、二つが挙げられる。
蓄積するときの割合を変える。
スレッショルド自体を変える
System design
Our project uses two classes of bacteria: senders and receivers.Senders produce signaling molecules, and receivers are activated only after a particular concentration of this molecule is reached.The communication using non-endogenous molecules is less sensitive,and it requires higher signal concentration to take effect.This results in slower activation of receivers.
About Quorum Sensing
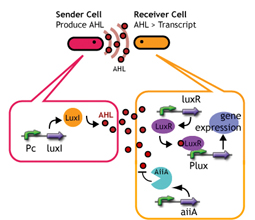
Quorum sensing is a cell-to-cell signaling action of bacteria. Species of Gram-Negative signaling transfer molecules (so-called autoinducer) is a series of acyl homoserine lactone (AHL). The signals are synthesized by a synthase protein(LuxI-type proteins) and once they have reached a threshold concentration,they bound to a transcriptional regulatory protein(LuxR-type proteins) to induce expression of target genes.
More about Quorum Sensing
- [http://parts.mit.edu/registry/index.php/Featured_Parts:Cell-Cell-Signaling Cell-Cell-Signaling]
- [http://www.che.caltech.edu/groups/fha/quorum.html About Quorum sensing]
Controlling the time of a cell-to-cell signaling action
Communication using non-endogenous molecules is less sensitive, and requires a higher signal concentration to take effect.This results in slower activation of receivers.
シグナル分子に対する応答速度を変えるためには3つの方法が考えられる。
- シグナル分子を作るSenderを変える
- シグナルを受け取るReceiverを変える
- シグナル分子自体を分解して、邪魔をする
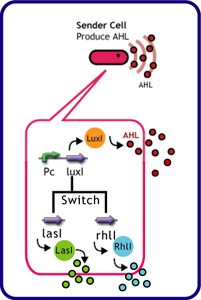
Signal Molecule Sender
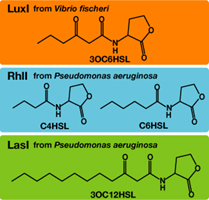
English:Each species has their own LuxI-type proteins,which synthesize their specific autoinducers,AHLs.AHLs produced by different LuxI-type proteins differ only in the length of the acyl-chain moiety and substitution at position C-3.Vibrio fischeri has LuxI,which synthesize 3OC6HSL.Pseudomonas aeruginosa has RhlI and LasI,which synthesize C4HSL、C6HSL,respectively.LuxR,which is original for Vibrio fischeri,is activated by its cognate autoinducer,3OC6HSL.However,LuxR is also activated by non-endogenous molecules,C4HSL,C6HSL,and 3OC12HSL.Activation by non-endogenous molecules requires a higher signal concentration[http://partsregistry.org/Part:BBa_F2620:Specificity (2)].This results in slower activation of receivers,when AHL concentration is increasing.
日本語:異なる生物はそれぞれに異なるLuxIタイプのタンパク質を持ち、アシル鎖の長さ、あるいはC-3位の置換基が異なる種類のAHLを合成する。Vibrio fischeriはLuxIにより3OC6HSLを、Pseudomonas aeruginosaはRhlIにより、C4HSL、C6HSLを、LasIにより3OC12HSLを合成する(Fig.4).AHLを受け取り応答するLuxRタンパク質はVibrio fischeri由来であり、低濃度の3OC6HSLに対して(~5nM)、応答する。しかし、他種生物由来のAHLに対しても応答することが知られており、より高い濃度のAHLが必要となる[http://partsregistry.org/Part:BBa_F2620:Specificity (2)].AHLがゆっくり溜まっていく時、LuxRは3OC6HSLに対して最も早く応答し、他のAHLに対してはそれよりも遅く応答する。
(冨永)
Sender experiments detail
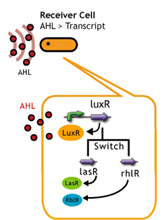
Signal Molecule Receiver
English:
日本語:AHLを合成するSenderだけではなく、AHLを受け取る側のReceiverを変えれば、その応答時間を変えることができる。そこで私たちは、以下のいくつかの方法を考えた。
- 一種類のSender(AHL<--LuxI)に対して、由来生物の異なるレシーバタンパク質でそれを受信する.
- レシーバータンパク質であるLuxRに変異を入れることで、AHLに対する応答感度を上下させること.
- レシーバーのコピーナンバーを変える.
(福冨、小林)
Quorum-Sensing Cross-talk
English:
日本語:
Copy nomber of Receiver
レシーバーのコピーナンバーを減らすことで、thresholdに達するまでの時間を短縮する。
LuxR/Plux mutants show
私たちはmutated Receiverを用いることで、AHL感受性の違う2種類(WTと変異体)のレシーバーを用意し、delay-timeのバリエーションを増やした。
- a greater response to 3OC6HSL[http://authors.library.caltech.edu/5553/ (3)]
- a increase in sensitivity to 3OC12HSL[http://mic.sgmjournals.org/cgi/content/abstract/151/11/3589 (4)].
Receiver experiments detail
Signal Molecule Quencher
AiiAによるAHL分解を同時に起こすことで、AHL供給速度を遅くする.
- AHL reporter with aiiA
- Express LuxR and aiiA constantly. AiiA degrades AHL as signaling molecule. Express GFP when the AHL concentration exceed the capacity of aiiA.
- This enables the delay of the activation time of receiver.
AiiA experiments detail
Demo Experiments
実験内容とdataかるく
Demo experiments detail
Conclusion
けつろん
Future Work
references
- [http://www3.interscience.wiley.com/journal/119124142/abstract M.K Winson et al.:Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing.FEMS Microbiology Letters 163 (1998) 185-192]
- [http://partsregistry.org/Part:BBa_F2620:Specificity BBa_F2620:Specificity]
- [http://authors.library.caltech.edu/5553/ C. H. Collins.et al.:Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones.Mol.Microbiol.2005.55(3).712–723]
- [http://mic.sgmjournals.org/cgi/content/abstract/151/11/3589 B. Koch. et al.:The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors.Microbiology 151 (2005),3589-3602]
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
 "
"