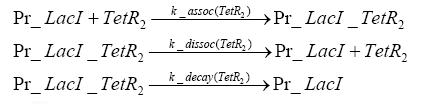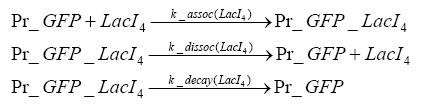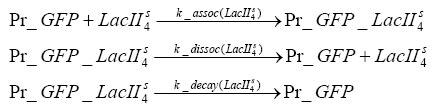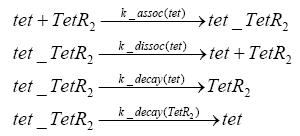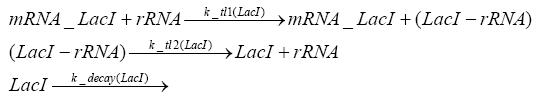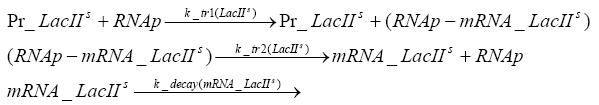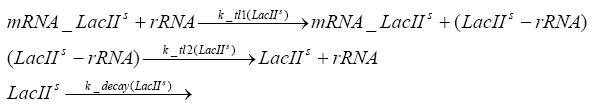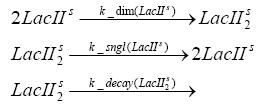Team:ETH Zurich/Modeling/Switch Circuit
From 2008.igem.org
(→diffusion of IPTG) |
(→diffusion of IPTG) |
||
| Line 15: | Line 15: | ||
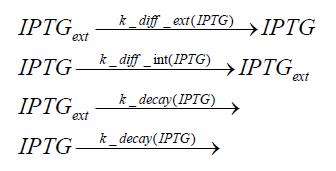
=== diffusion of IPTG === | === diffusion of IPTG === | ||
| - | The circuit is induced by IPTG which is added into the medium and diffuses into the cell and from the cell the IPTG can diffuse back into the medium. Moreover IPTG also degrades slowly inside the cell and also outside where it is additionally washed away in case if we run the whole system inside a chemostat. | + | The circuit is induced by IPTG which is added into the medium and diffuses into the cell and from the cell the IPTG can diffuse back into the medium. Moreover IPTG also degrades slowly inside the cell and also outside where it is additionally washed away in case if we run the whole system inside a chemostat.<br> |
<html><img src="https://static.igem.org/mediawiki/2008/6/62/Circuit_iptg_diff.JPG" alt="Grafik" width="30%"></img></html> | <html><img src="https://static.igem.org/mediawiki/2008/6/62/Circuit_iptg_diff.JPG" alt="Grafik" width="30%"></img></html> | ||
[[Image:diffusion_IPTG.JPG|left width=30%]] | [[Image:diffusion_IPTG.JPG|left width=30%]] | ||
Revision as of 18:53, 27 October 2008
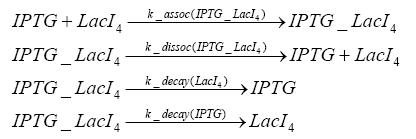
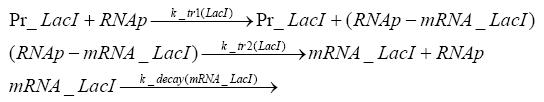
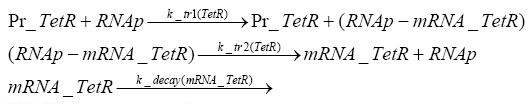
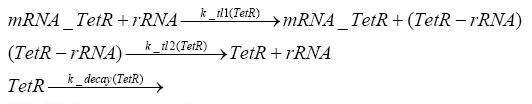
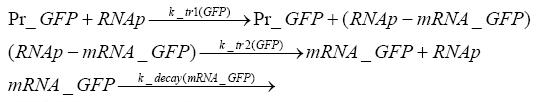
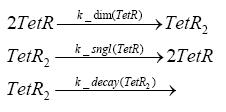
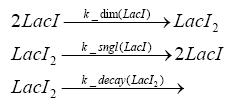
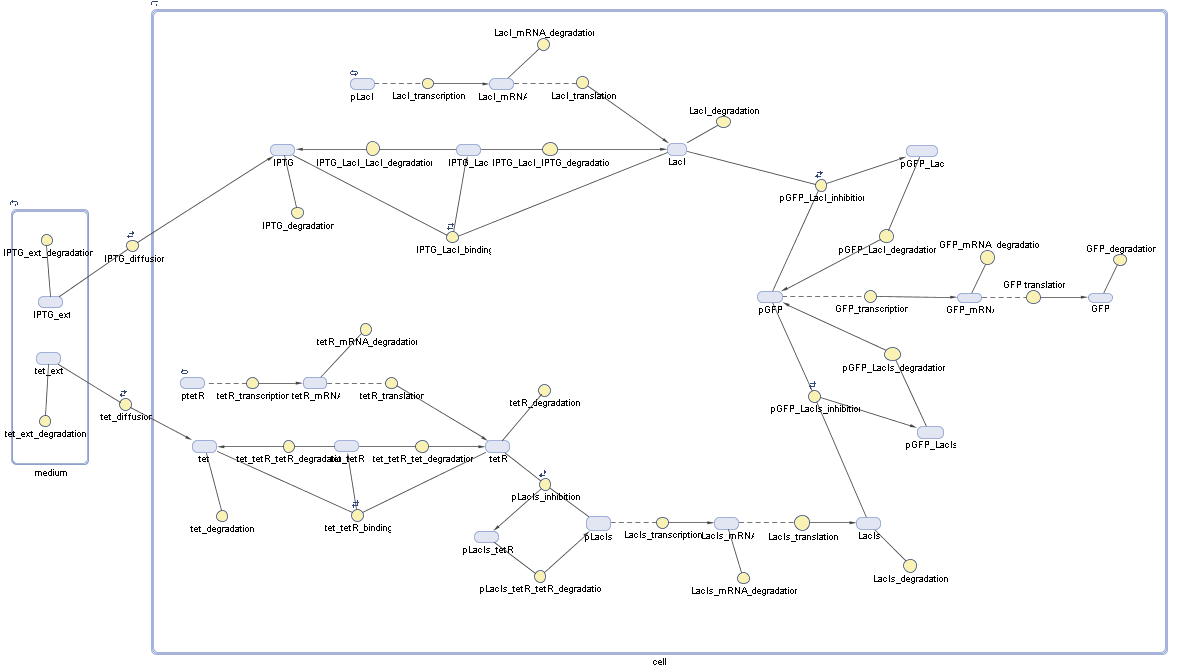
Switch CircuitDetailed Modeldiffusion of IPTGThe circuit is induced by IPTG which is added into the medium and diffuses into the cell and from the cell the IPTG can diffuse back into the medium. Moreover IPTG also degrades slowly inside the cell and also outside where it is additionally washed away in case if we run the whole system inside a chemostat. diffusion of tetbinding of tetR to LacI-promotor and LacIIs-promotorbinding of LacI and LacIIs to GFP-promotorbinding of tet to tetRbinding of IPTG to LacItranscription and translation of LacItranscription and translation of LacIIstranscription and translation of tetRtranscription and translation of GFPdimerization of tetRdimerization and tetramerization of LacI and LacIIsImplementationThe model of the switch circuit has been implemented using the Simbiology Toolbox in MATLAB. For the sake of simplicity, the effects of dimerization and dimerization/tetramerization of tetR and LacI/LacIIs have been neglected. So we did with the additional steps involving the RNApolymerase in the transcription of proteins and the ribosomes in the translation of proteins. SimulationIn order to get some useful results out of this model, we ran deterministic and stochastic simulations. Results and DiscussionParametersIn this section you can find all the parameters used in the simulation.
References(1) "Spatiotemporal control of gene expression with pulse-generating networks", Basu et al., PNAS, 2004 (2) "Genetic circuit building blocks for cellular computation, communications, and signal processing", Weiss et al., Natural Computing, 2003 (3) "Predicting stochastic gene expression dynamics in single cells", Mettetal et al., PNAS, 2006 (4) "Engineered gene circuits", Hasty et al., Nature, 2002 |
 "
"