Team:Imperial College/Major Results/B.subtilis
From 2008.igem.org
m |
m |
||
| Line 1: | Line 1: | ||
{{Imperial/StartPage2}} | {{Imperial/StartPage2}} | ||
<br> | <br> | ||
| - | {{Imperial/Box2|Working With ''B.subtilis''|Throughout this summer we worked with ''B.subtilis'' as a chassis. Although this chassis has great potential it is not commonly used and very few ''B.subtilis'' biobricks are available. We aimed to increase the accessibility of ''B.subtilis'' as a chassis. The major results of our work is summarised on this page. | + | {{Imperial/Box2|Working With ''B. subtilis''|Throughout this summer we worked with ''B. subtilis'' as a chassis. Although this chassis has great potential it is not commonly used and very few ''B. subtilis'' biobricks are available. We aimed to increase the accessibility of ''B. subtilis'' as a chassis. The major results of our work is summarised on this page. |
}} | }} | ||
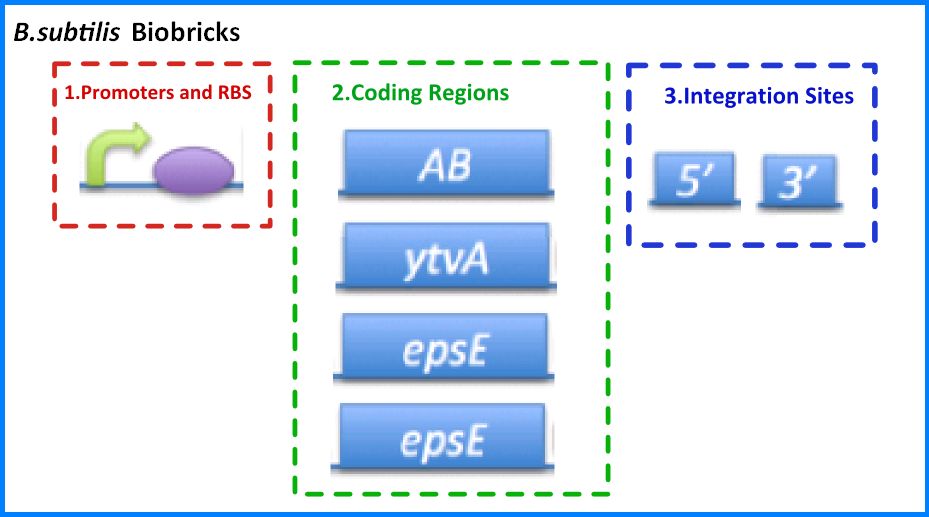
| - | {{Imperial/Box1|''B.subtilis'' Biobricks|In order to pursue our project we had to design and build a variety of biobricks parts for ''B.subtilis''. These can be split up into the parts: | + | {{Imperial/Box1|''B. subtilis'' Biobricks|In order to pursue our project we had to design and build a variety of biobricks parts for ''B. subtilis''. These can be split up into the parts: |
*'''Promoter and Ribosome Binding Sites (RBS)''' - These promoter and RBS combinations include a number of inducible and constitutive promoters combined with weak and strong ribosome binding sites. | *'''Promoter and Ribosome Binding Sites (RBS)''' - These promoter and RBS combinations include a number of inducible and constitutive promoters combined with weak and strong ribosome binding sites. | ||
*'''Coding Regions''' - These parts include specific coding regions concerned with our project such as biomaterials and in addition more general coding regions such as antibiotic resistance genes that can be used for selection markers. | *'''Coding Regions''' - These parts include specific coding regions concerned with our project such as biomaterials and in addition more general coding regions such as antibiotic resistance genes that can be used for selection markers. | ||
| - | *'''Integration Sites''' - As an alternative to maintaining genetic devices on extra-chromosomal plasmids we investigated the construction of parts to allow integration of genetic devices into the ''B.subtilis'' chromosome. | + | *'''Integration Sites''' - As an alternative to maintaining genetic devices on extra-chromosomal plasmids we investigated the construction of parts to allow integration of genetic devices into the ''B. subtilis'' chromosome. |
|[[Image:B.subtilis parts.PNG|right|300px]] | |[[Image:B.subtilis parts.PNG|right|300px]] | ||
}} | }} | ||
| - | {{Imperial/Box1|Transformation of ''B.subtilis''|In order to expand the use of ''B.subtilis'' as a potential chassis we investigated the different protocols used for the transformation of ''B.subtilis''. ''B.subtilis'' is used as the model for gram positive bacterium and have been extensively studied. A variety of protocols have been developed for the transformation of ''B.subtilis''. We investigated two different protocols, one relying upon natural competence of stressed ''B.subtilis'' and the using electroporation. The protocols and results were both fully documented to further the use of ''B.subtilis'' as a chassis. | + | {{Imperial/Box1|Transformation of ''B. subtilis''|In order to expand the use of ''B. subtilis'' as a potential chassis we investigated the different protocols used for the transformation of ''B. subtilis''. ''B. subtilis'' is used as the model for gram positive bacterium and have been extensively studied. A variety of protocols have been developed for the transformation of ''B. subtilis''. We investigated two different protocols, one relying upon natural competence of stressed ''B. subtilis'' and the using electroporation. The protocols and results were both fully documented to further the use of ''B. subtilis'' as a chassis. |
*The protocols can be found here -[https://2008.igem.org/Team:Imperial_College/Transformation Protocol 1] and [https://2008.igem.org/Team:Imperial_College/Transformation_2 Protocol 2] | *The protocols can be found here -[https://2008.igem.org/Team:Imperial_College/Transformation Protocol 1] and [https://2008.igem.org/Team:Imperial_College/Transformation_2 Protocol 2] | ||
*The summary of results can be found here -[https://2008.igem.org/Team:Imperial_College/Transformation_Results Summary of results] | *The summary of results can be found here -[https://2008.igem.org/Team:Imperial_College/Transformation_Results Summary of results] | ||
| Line 16: | Line 16: | ||
{{Imperial/Box1|Chassis Characterisation| | {{Imperial/Box1|Chassis Characterisation| | ||
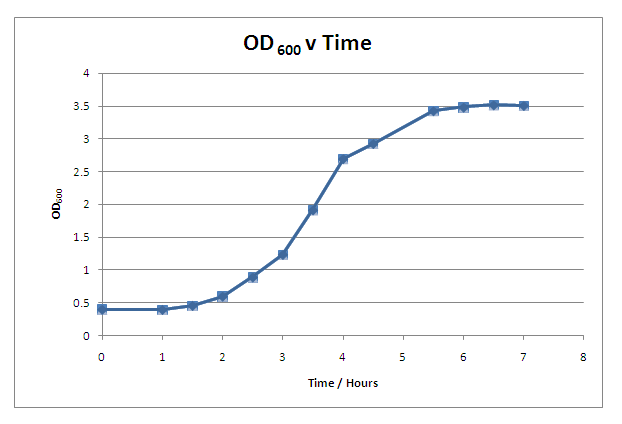
'''Growth Curve'''<br> | '''Growth Curve'''<br> | ||
| - | We characterised the growth rate of ''B.subtilis'' in LB media at 37<sup>o</sup>C. This curve provides useful information when inducing protein synthesis in ''B.subtilis'' and when characterising parts such as promoters. In addition to collecting this growth curve information, we modeled the growth rate of ''B.subtilis'' which can be found by [https://2008.igem.org/Team:Imperial_College/Growth_Curve clicking this link].| | + | We characterised the growth rate of ''B. subtilis'' in LB media at 37<sup>o</sup>C. This curve provides useful information when inducing protein synthesis in ''B. subtilis'' and when characterising parts such as promoters. In addition to collecting this growth curve information, we modeled the growth rate of ''B. subtilis'' which can be found by [https://2008.igem.org/Team:Imperial_College/Growth_Curve clicking this link].| |
[[Image:OD600 v Time.PNG|center|300px]] | [[Image:OD600 v Time.PNG|center|300px]] | ||
}} | }} | ||
Revision as of 11:38, 28 October 2008
|
||||||||||||||||
 "
"