Team:Newcastle University/Parts Repository
From 2008.igem.org
| Line 10: | Line 10: | ||
Computers do not know any biology. Many of the other projects involve manipulating data without any need to understand its biological meaning. This module is responsible for capturing biological knowledge and how it relates to that data. So the parts that make up a biological system need to be described in a computationally accessible manner. This system will model the parts that make up the biological circuits. Efforts to define parts are already underway in the BioBricks community, and this project will build upon these. | Computers do not know any biology. Many of the other projects involve manipulating data without any need to understand its biological meaning. This module is responsible for capturing biological knowledge and how it relates to that data. So the parts that make up a biological system need to be described in a computationally accessible manner. This system will model the parts that make up the biological circuits. Efforts to define parts are already underway in the BioBricks community, and this project will build upon these. | ||
| + | |||
| + | The requirement for a repository of models to complement defined parts and assist synthetic design has not been neglected (Le Novere ''et al.'', 2006; Rodrigo, ''et al.'', 2007; Rouilly ''et al.'', 2007). However, to date these needs have not been met by a suitable repository of part models. In January 2007, Rouilly ''et al.'' announced that creating a repository of CellML models to correspond to the BioBricks parts registry was both useful and achievable. Since this declaration, a registry of biological models has been created yet populated by barely more than 10 models, many of which are not validated (Registry of Standard Biological Models, 2007). CellML models have been created for generic parts such as promoters, ribosome-binding sites, mRNA, proteins and oscillators (Registry of Standard Biological Models, 2007). | ||
| + | |||
| + | |||
</div> | </div> | ||
| Line 148: | Line 152: | ||
<html><a name="java-programme"></html> | <html><a name="java-programme"></html> | ||
==== iGEM Java programme ==== | ==== iGEM Java programme ==== | ||
| + | |||
| + | The Java programme requires a level of communication to enable access of the PR to query information. JDBC(TM) was used to provide a connection between the Java code and the repository stored as a Microsoft Access database. Firstly a JDBC-ODBC bridge was installed, then using the ODBC data source administrator, a Microsoft Access driver was set up to access the PR. The sun JDBC driver was used (sun.jdbc.odbc.JdbcOdbcDriver). A Java programme was written to access data from the repository. Once an interface and its relative classes were created, a temporary interface was designed for the interaction of the parts repository and evolutionary algorithm. | ||
This is an interface that allows easy access to the items stored in the PR database without needing knowledge of the database structure. It is written in Java to access via webservices. The Javadoc is available. | This is an interface that allows easy access to the items stored in the PR database without needing knowledge of the database structure. It is written in Java to access via webservices. The Javadoc is available. | ||
Revision as of 17:39, 28 October 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Original Aims >> Parts Repository
Parts Repository
Aims and Objectives
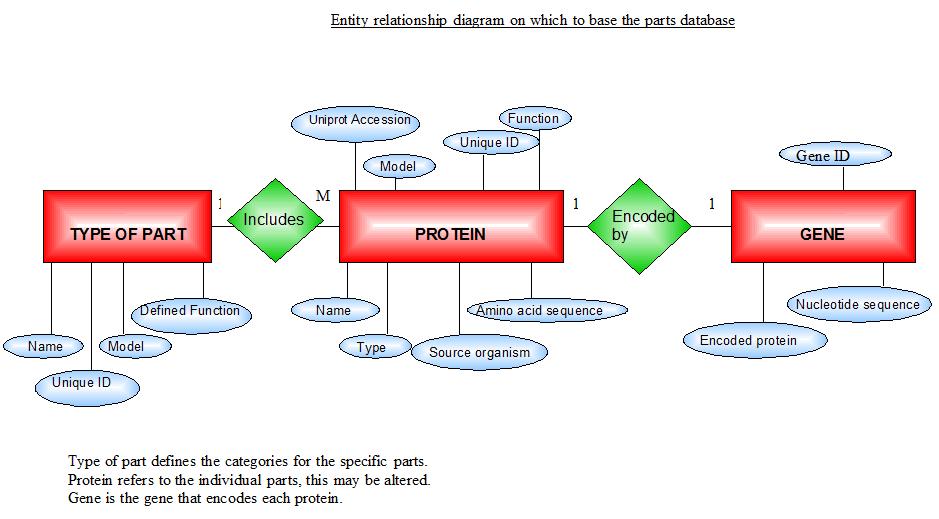
To develop a system that will define the kinds of parts that make up a biological circuit and their behaviour.
Computers do not know any biology. Many of the other projects involve manipulating data without any need to understand its biological meaning. This module is responsible for capturing biological knowledge and how it relates to that data. So the parts that make up a biological system need to be described in a computationally accessible manner. This system will model the parts that make up the biological circuits. Efforts to define parts are already underway in the BioBricks community, and this project will build upon these.
The requirement for a repository of models to complement defined parts and assist synthetic design has not been neglected (Le Novere et al., 2006; Rodrigo, et al., 2007; Rouilly et al., 2007). However, to date these needs have not been met by a suitable repository of part models. In January 2007, Rouilly et al. announced that creating a repository of CellML models to correspond to the BioBricks parts registry was both useful and achievable. Since this declaration, a registry of biological models has been created yet populated by barely more than 10 models, many of which are not validated (Registry of Standard Biological Models, 2007). CellML models have been created for generic parts such as promoters, ribosome-binding sites, mRNA, proteins and oscillators (Registry of Standard Biological Models, 2007).
The objectives of this project are:
- To develop a database that stores clear definitions of the components of a genetic circuit, including:
- sequences
- important domains
- abstract model(s)
- the ontology of component types
- provenance back to e.g. BioBrick of origin
- To develop an API that will provide access to the repository in a standard, easily usable manner
Outcomes
The work for the Parts Repository (PR) was concluded on 1 Sept 2008.
Parts Repository
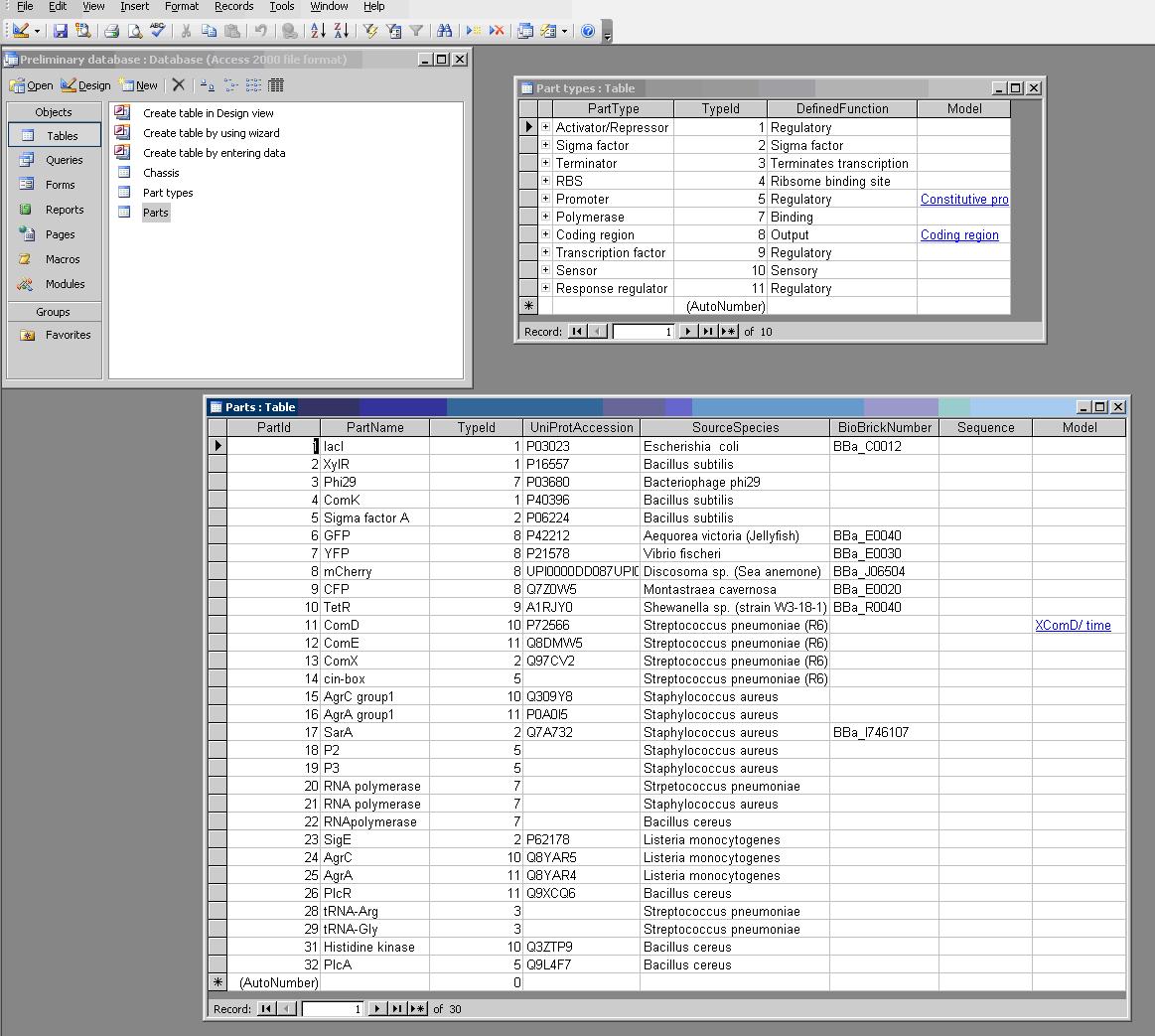
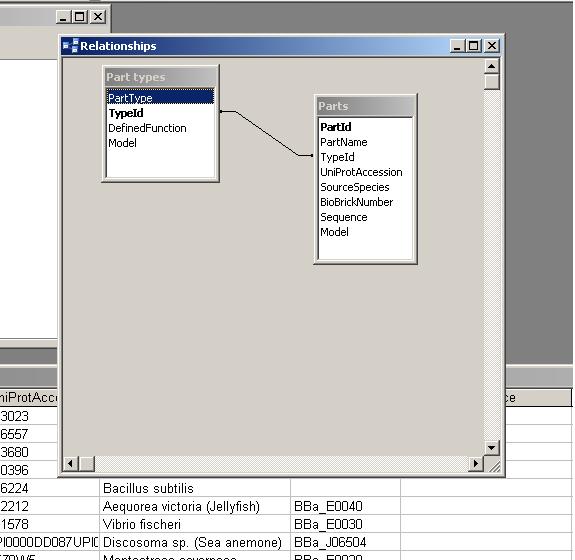
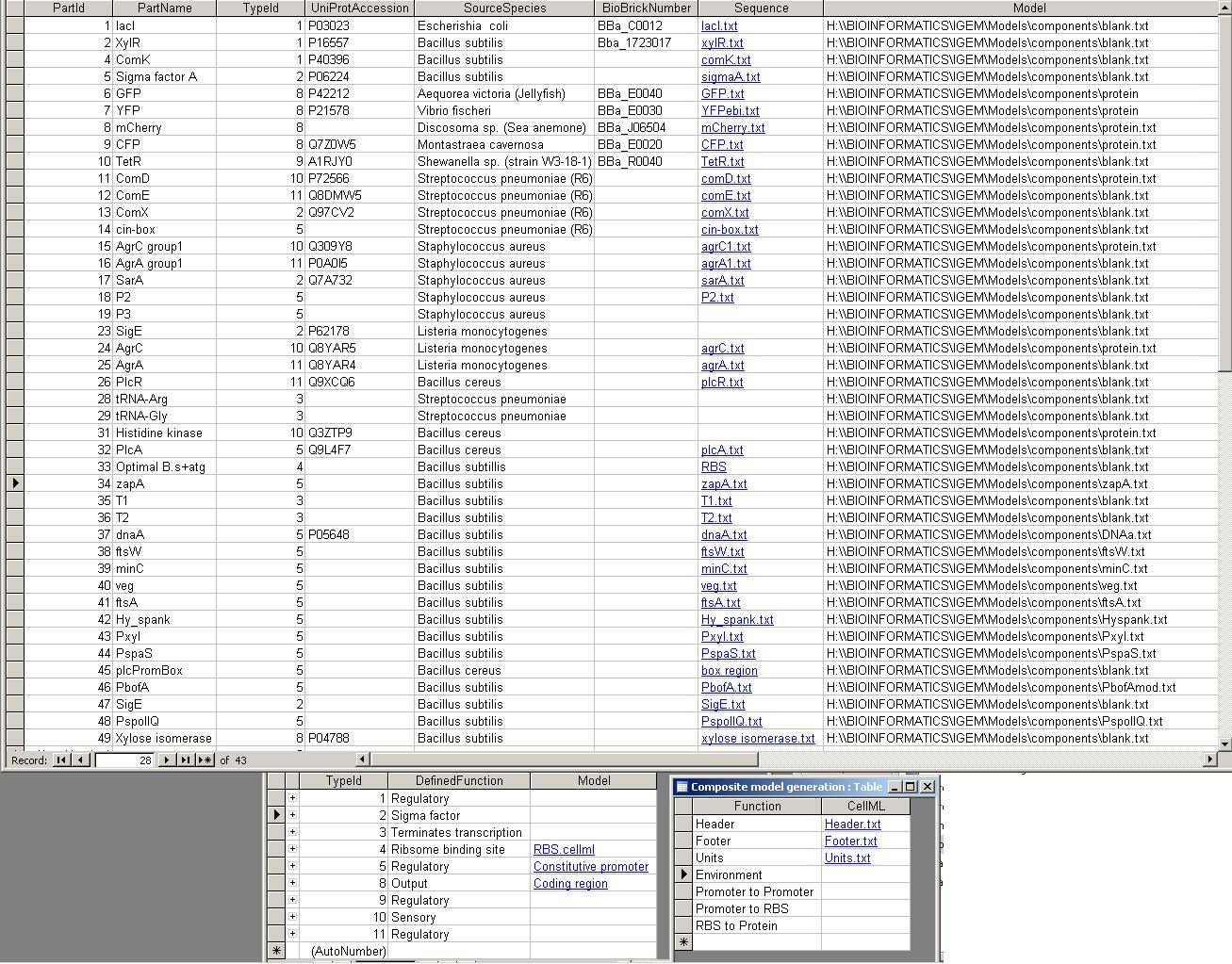
The database repository of genetic parts for bottom-up modelling of synthetic biology. Implemented as a Microsoft Access database.
The PR attempts to completely define a "part", which can be any genetically defined construct. These can include BioBrick parts and devices, as well as independently defined objects like the 2-part quorum sensing systems used in the BugBuster project.
Part Sequences
The nucleotide sequences for the parts used in the BugBuster project, to further define the function of a part in the part repository.
CellML models
The behaviour of each part was defined by a [http://www.cellml.org/ CellML model].
Some of the models are complete, and others are incomplete. It is suggested to compile the models in [http://cor.physiol.ox.ac.uk/ COR] and to run them in [http://www.cellml.org/tools/pcenv/ PCEnv].
There are also some components, which are the fragments of CellML code that are associated with each part, and fragments to create complete models from these. They appear as they are stored in the parts repository for access by the iGEM project as a whole.
Download: File:Newcastle-igem2008-CellML-models.zip contains:
- Alt.promoter.constitutive.cellml
- Alt.promoter.constitutiveImport.cellml
- Alt.promoter.inductive.cellml
- Alt.promoter.inductive3.cellml
- Alt.promoter.respressive.cellml
- Alt.protein.cellml
- Alt.RBS.cellml
- Bacillus RBS.cellml
- Basic promoter 2.cellml
- Basic promoter 3.cellml
- Basic Promoter.cellml
- Check comp.cellml
- CodingRegion.cellml
- ComD(NoInducSynthWithComEP).cellml
- ComD.cellml
- ComDAGAIN.cellml
- composite2.cellml
- CompositeModel.cellml
- constitutive promoter.cellml
- DNAa.cellml
- EncodedProtein.cellml
- example tut3main.cellml
- example tut3sub..cellml
- FluxComEP.cellml
- ftsA.cellml
- ftsW.cellml
- HySpank.cellml
- InitialRBS.cellml
- minC.cellml
- NewComD.cellml
- PbofA.cellml
- PcotA.cellml
- Protein.cellml
- proteinCoding.cellml
- PspaS.cellml
- PspollQ.cellml
- Pxyl.cellml
- ResponseRegulator.cellml
- Sensor.cellml
- Tanscription factor.cellml
- To join the components together.doc
- veg.cellml
- XComDP.cellml
- zapA.cellml
File:Newcastle-igem2008-CellML-components.zip contains:
- AgrA.txt
- AgrC.txt
- Bind Components.txt
- CFP.txt
- Cin-box.txt
- ComD.txt
- ComE.txt
- ConstitutivePromoter.txt
- DNAa.txt
- Environment.txt
- Essentials.txt
- Footer.txt
- ftsA.txt
- ftsW.txt
- GFP.txt
- Header.txt
- HySpank.txt
- InducablePromoter.txt
- mCherry.txt
- minC.txt
- OptimalRBS.txt
- P2.txt
- P3.txt
- PbofA.txt
- PbofAmod.txt
- PcotA.txt
- PlcA.txt
- PromoterToRBS.txt
- Protein.txt
- PspaS.txt
- PspollQ.txt
- Pxyl.txt
- RBS.txt
- RBSToCodingRegion.txt
- RepressablePromoter.txt
- ResponseRegulator.txt
- Sensor.txt
- Units.txt
- veg.txt
- XyloseIsomerase.txt
- YFP.txt
- YocF.txt
- YocG.txt
- zapA.txt
iGEM Java programme
The Java programme requires a level of communication to enable access of the PR to query information. JDBC(TM) was used to provide a connection between the Java code and the repository stored as a Microsoft Access database. Firstly a JDBC-ODBC bridge was installed, then using the ODBC data source administrator, a Microsoft Access driver was set up to access the PR. The sun JDBC driver was used (sun.jdbc.odbc.JdbcOdbcDriver). A Java programme was written to access data from the repository. Once an interface and its relative classes were created, a temporary interface was designed for the interaction of the parts repository and evolutionary algorithm.
This is an interface that allows easy access to the items stored in the PR database without needing knowledge of the database structure. It is written in Java to access via webservices. The Javadoc is available.
Further Reading
Contributors
Lead: Megan Aylward
</div>
 "
"