Data Retrieval and Storage
From 2008.igem.org
(→UniProt) |
(→UniProt) |
||
| Line 37: | Line 37: | ||
<div align=justify> | <div align=justify> | ||
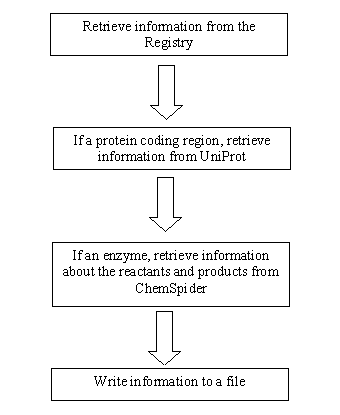
| - | IIf a protein makes up one of the parts retrieved from the iGEM database, that result is sent to UniProt. UniProt is a large database of proteins and enzymes. This database can be queried by a Blast algorithm, which is a very powerful programming tool. When inputting the DNA or amino acid sequence, UniProt gives results that are closest to the initial search. Besides giving the name of the protein searched, UniProt will give the reagents from the reaction that the protein catalyzes. All this information is useful for EvoGEM and is stored in a local database. Visit | + | IIf a protein makes up one of the parts retrieved from the iGEM database, that result is sent to UniProt. UniProt is a large database of proteins and enzymes. This database can be queried by a Blast algorithm, which is a very powerful programming tool. When inputting the DNA or amino acid sequence, UniProt gives results that are closest to the initial search. Besides giving the name of the protein searched, UniProt will give the reagents from the reaction that the protein catalyzes. All this information is useful for EvoGEM and is stored in a local database. Visit [http://www.uniprot.com Uniprot] to see this database. </div> |
<br style="clear:both"/> | <br style="clear:both"/> | ||
Revision as of 23:35, 28 October 2008

|
| Home | The Team | The Project | Modeling | Notebook |
|---|
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
Contents |
Perl
- If they were enzymes, what reactions were they catalyzing? - If they were molecules, what were the molecular structures or other synonyms for these compounds?
The answers to these questions would allow EvoGEM to distinguish between different compounds better. How do we accomplish this, though? By creating a Perl script! Perl is a programming language that is powerful in text processing facilities. Since it effectively uses string matching, it is an ideal language for searching text and manipulating text files, which is exactly what we need for retrieving and expanding EvoGEM's local database.
UniProt
ChemSpider
After results are gone through UniProt, if there are further molecules that are involved in the reaction that are not proteins, the search goes to ChemSpider. This large database is much like UniProt except that it is for chemistry. Searching and querying in ChemSpider is quite simple as things can be queried using synonyms of molecules. This makes it a very useful tool. After a molecule is queried, ChemSpider will produce information about the molecule such as synonyms and SMILES, which is a simplified molecular input line entry specification. As useful as this information can be, the reason for coming for this database is to get something that is machine readable and can be used for comparisons of metabolic pathways. What is this machine readable format? This machine readable format is known as the IUPAC International Chemical Identifier (InChI). This InChi is a unique "fingerprint" of the molecule that is not ambiguous like SMILES and is supplied only by IUPAC. An example of an InChi would look like this:
1/C6H8O6/c7-1-2(8)5-3(9)4(10)6(11)12-5/h2,5,7-8,10-11H,1H2/t2-,5+/m0/s1
To see this database, go here: [http://www.chemspider.com ChemSpider]
The Algorithm
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
| Home | The Team | The Project | Modeling | Notebook |
|---|
 "
"