Team:Calgary Software/Project
From 2008.igem.org
(→Modeling) |
(→Introduction) |
||
| Line 29: | Line 29: | ||
This design uses the paradigm of evolution to select efficient designs and produce a product or output that is generated independently by the system. EvoGEM uses the strategies of genetics and evolution to simulate an environment inside of a prokaryotic cell. This entails various events and structures that are present inside of the organism, such as: | This design uses the paradigm of evolution to select efficient designs and produce a product or output that is generated independently by the system. EvoGEM uses the strategies of genetics and evolution to simulate an environment inside of a prokaryotic cell. This entails various events and structures that are present inside of the organism, such as: | ||
| - | + | * RNA polymerase | |
| - | + | * messenger RNA | |
| - | + | * ribosomes | |
| - | + | * transcription and translation. | |
The program assembles generations from a selection of parts - retrieved from the iGEM registry - and creates genetic circuits. The circuits proliferate in each generation, and the best possible combination of parts is selected. Eventually, the best circuit is obtained from the system and a desired functionality is returned. The powerful agent-based logic allows for minimal assumptions about the behavior of the system, when coupled with the empirically proven evolutionary design, creates a superb system that is able to both simulate and develop iGEM circuits. </div> | The program assembles generations from a selection of parts - retrieved from the iGEM registry - and creates genetic circuits. The circuits proliferate in each generation, and the best possible combination of parts is selected. Eventually, the best circuit is obtained from the system and a desired functionality is returned. The powerful agent-based logic allows for minimal assumptions about the behavior of the system, when coupled with the empirically proven evolutionary design, creates a superb system that is able to both simulate and develop iGEM circuits. </div> | ||
| Line 40: | Line 40: | ||
EWe presented EvoGEM during the 2007 iGEM Jamboree, where it sparked a lot of interest amongst the different teams. This summer, our team has expanded EvoGEM by: | EWe presented EvoGEM during the 2007 iGEM Jamboree, where it sparked a lot of interest amongst the different teams. This summer, our team has expanded EvoGEM by: | ||
| - | + | * improving EvoGEM's fitness function. | |
| - | + | * introducing more complex pattern recognition. | |
| - | + | * testing the system under a much larger search space. | |
Our goal was to create a system sophisticated enough to rebuild working designs from previous years' teams' projects, as well as intelligent enough to simulate successes and failures of working and non-working systems, respectively. | Our goal was to create a system sophisticated enough to rebuild working designs from previous years' teams' projects, as well as intelligent enough to simulate successes and failures of working and non-working systems, respectively. | ||
We achieved our goals by: | We achieved our goals by: | ||
| - | + | * building Perl scripts that support EvoGEM's requirements of a flat file registry | |
| - | + | * creating an Objective-C based graphical user interface (GUI) in order to make the software-user interaction easy | |
| - | + | * developing the EvoGEM code to include the behaviors specified before, and create a simulation of the processes in the cell such as transcription and translation. | |
</p> | </p> | ||
Revision as of 23:46, 28 October 2008

|
| Home | The Team | The Project | Modeling | Notebook |
|---|
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
Contents |
Introduction
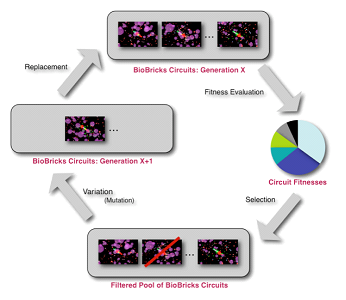
We use evolutionary and genetic strategies, which are useful modeling methods, especially when coupled with agent-based designs. We created EvoGEM, a design that is based on the registry of genetic parts provided by the iGEM competition.
This design uses the paradigm of evolution to select efficient designs and produce a product or output that is generated independently by the system. EvoGEM uses the strategies of genetics and evolution to simulate an environment inside of a prokaryotic cell. This entails various events and structures that are present inside of the organism, such as:
- RNA polymerase
- messenger RNA
- ribosomes
- transcription and translation.
EWe presented EvoGEM during the 2007 iGEM Jamboree, where it sparked a lot of interest amongst the different teams. This summer, our team has expanded EvoGEM by:
- improving EvoGEM's fitness function.
- introducing more complex pattern recognition.
- testing the system under a much larger search space.
- building Perl scripts that support EvoGEM's requirements of a flat file registry
- creating an Objective-C based graphical user interface (GUI) in order to make the software-user interaction easy
- developing the EvoGEM code to include the behaviors specified before, and create a simulation of the processes in the cell such as transcription and translation.
Evolutionary Algorithm
Evolution involves the changes of inherited traits in a population for successive generations. Each generation carries genetic information, expressing certain characteristics. Mutation enables manipulation of these traits as well as genetic recombination. Evolution is a result of heritable traits becoming more prevalent or rare. Agent-based modeling is a computational method that replicates the behavior and interaction of individual components of a network such that their overall effect on the system can be observed. This involves many different aspects, including: - game theory - evolutionary programming - complex systems - emergence
Multiple agents are simulated in an environment to simulate and hypothesize the actions of complex phenomena. .
Data Retrieval and Storage
- its type. - its function. - whether it codes for a protein. - how well the parts work. If the part coded for a protein, we retrieved its DNA sequence. We, then, found the amino acid sequence (using the BLAST algorithm) from UniProt, which is a large database of proteins. If the protein catalyzed an associated prosthetic or a biochemical reaction, we retrieved additional information from ChemSpider - a chemical database - to find the data characterizing any compounds that the particular enzymatic protein catalyzes. Finally, we stored the data in a database during run-time of EvoGEM.
Modeling
Graphical User Interface
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
| Home | The Team | The Project | Modeling | Notebook |
|---|
 "
"