Team:Newcastle University/Workbench
From 2008.igem.org
| Line 22: | Line 22: | ||
*take in conditions for synthetic evolution and send models to the [[Team:Newcastle University/Evolutionary Algorithm|evolutionary algorithm]] | *take in conditions for synthetic evolution and send models to the [[Team:Newcastle University/Evolutionary Algorithm|evolutionary algorithm]] | ||
*output the sequence in a standard format | *output the sequence in a standard format | ||
| + | <html><a name="user-stories"></html> | ||
| + | ===User Stories=== | ||
| + | ====Parts Repository==== | ||
| + | # The program must display a list of all available parts. The program requests a hierarchical list of all part names, part types, and their part ids from the Parts Repository (PR). Upon receiving the list, the program sorts the parts into different categories based on the part type, chooses a display icon for each type, and presents the list of part names with their part type icons to the user. | ||
| + | # The user wishes to change the type of chassis. The program retrieves a list of all chassis names and chassis ids from the PR and presents it to the user. | ||
| + | # The program must display a list of all types of parts. It requests a list of all part types and their part type ids from the PR. Then it displays the list of part names to the user. | ||
| + | # The program must display all of the information about a single part. It gives the part id to the PR and receives a full description of the part in return. It then displays the part information to the user. | ||
| + | # The program must save a CellML model of a part. It gives the part id to the PR and receives a segment of CellML in return. It then saves the CellML to file. | ||
| + | |||
| + | ====Constraints Repository==== | ||
| + | # The program must show all possible interactions to a particular part. It passes the part id to the Constraints Repository and receives a list of all possible interaction ids in return. It then retrieves the details of each interaction with the interaction id. It then shows the possible interactions to the user. | ||
| + | |||
| + | ====Evolutionary Algorithm==== | ||
| + | # The user finally wants to evolve her model in silico. The program collects the starting model, composed of a series of part ids, and the proposed behaviour in the form of a truth table, and gives it to the Evolutionary Algorithm (EA) for evolution. The program shows a “busy working” screen while the Evolutionary Algorithm computes the best model. When the EA finishes, the program receives the proposed best model(s) and displays them, along with their behaviour, to the user. | ||
==Outcome== | ==Outcome== | ||
| Line 30: | Line 44: | ||
The constraints repository aids in the building of this construct by not allowing parts that are not compatible to be dropped in place next to each other. Where parts are compatible with each other, constraint models are incorporated into the construct that define the interactions between these. | The constraints repository aids in the building of this construct by not allowing parts that are not compatible to be dropped in place next to each other. Where parts are compatible with each other, constraint models are incorporated into the construct that define the interactions between these. | ||
| + | |||
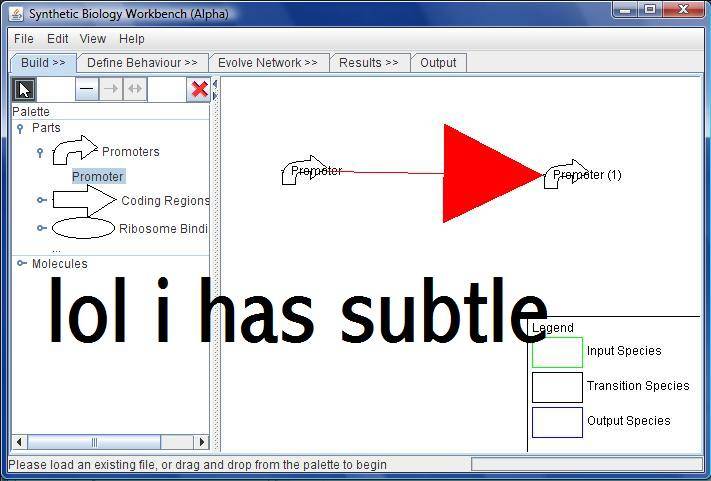
| + | [[Image:Subtle.jpg|thumb|right|DiS with a very subtle directed line, in a prototype version.]] | ||
The user can also map system inputs to system outputs that can be used by the EA. For example, the user could specify that in the presence of a certain set of quorum sensing peptides that a certain fluorescent protein would light up. The user can then run the EA through DiS, leading to the mutation of the hidden layer and the output of the hidden layer model that fulfils the output requirements the user has set. | The user can also map system inputs to system outputs that can be used by the EA. For example, the user could specify that in the presence of a certain set of quorum sensing peptides that a certain fluorescent protein would light up. The user can then run the EA through DiS, leading to the mutation of the hidden layer and the output of the hidden layer model that fulfils the output requirements the user has set. | ||
| + | |||
All of the different software components were supposed to communicate through DiS. Each team member was responsible for the construction of their component. The outcome of this was a working build, where the evolutionary algorithm could communicate directly with the parts repository, but not with DiS. The components were simply not ready to communicate with each other. Once some more work has been carried out on the various components of the overall project, DiS will be functional. | All of the different software components were supposed to communicate through DiS. Each team member was responsible for the construction of their component. The outcome of this was a working build, where the evolutionary algorithm could communicate directly with the parts repository, but not with DiS. The components were simply not ready to communicate with each other. Once some more work has been carried out on the various components of the overall project, DiS will be functional. | ||
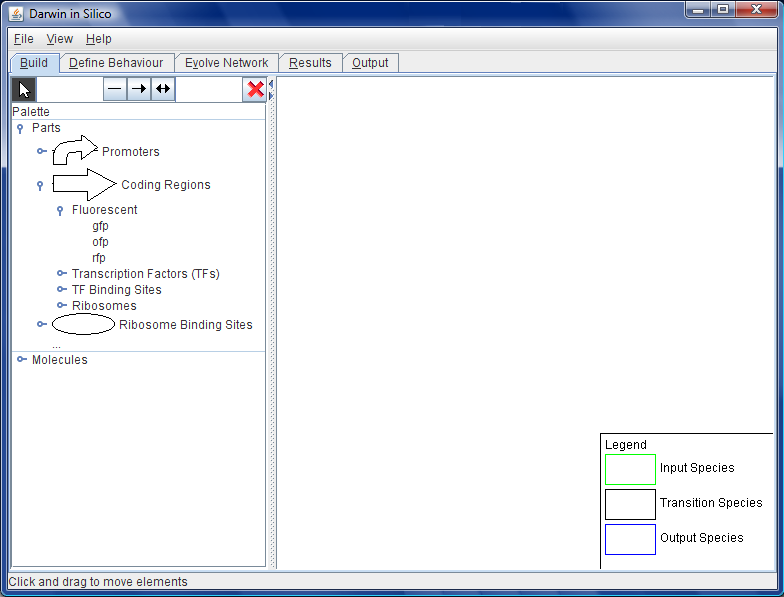
| + | [[Image:Wb-layout.jpg|thumb|left]] | ||
| + | |||
| + | ---- | ||
<html><table border="1" width="800"><tr><td valign="top"></html> | <html><table border="1" width="800"><tr><td valign="top"></html> | ||
| Line 48: | Line 68: | ||
[[Image:Wb-outputs.png|400px]] | [[Image:Wb-outputs.png|400px]] | ||
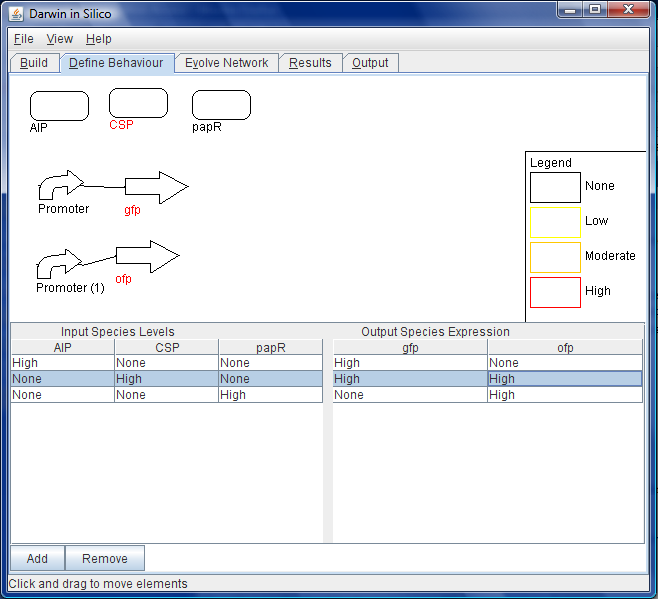
<br>In the ''Behaviour'' tab, the behaviour of the system is specified by a series of tables. In this image, when the input molecule "CSP" is present in high quantities, both GFP and OFP will also be expressed at high levels. | <br>In the ''Behaviour'' tab, the behaviour of the system is specified by a series of tables. In this image, when the input molecule "CSP" is present in high quantities, both GFP and OFP will also be expressed at high levels. | ||
| + | <html></td></tr><tr><td valign="top"></html> | ||
| + | [[Image:Wb-evolve.png|400px]] | ||
| + | <br> | ||
| + | <html></td><td valign="top"></html> | ||
| + | [[Image:Wb-results.png|400px]] | ||
| + | <br> | ||
<html></td></tr></table></html> | <html></td></tr></table></html> | ||
Revision as of 12:42, 29 October 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Workbench
Aims:
The proposed Synthetic Biology Workbench (cool name needed) will incorporate the functionality of a biological network modeling and simulation program along with spiffy new algorithms that can be used in modern synthetic biology. This will enable users to finetune the expression of models suitable for their purposes, particularly for such fun activities like iGEM competitions.
From a previously created repository of parts and devices, and using the (gentle) guidance of a set of assembly parameters, users will be able to create, edit and piece together biological networks at varying levels of complexity. Users will be able to piece together parts, adding ribosome binding sites, protein coding regions, terminators, etc. until they have exactly the device they desire.
Once the biological circuit is ready in principle, it can be put into practice. The user will be able to request the optimal biological circuit, composed from BioBricks from the repository.
Objectives:
- get a model
- load from SBML and/or CellML
- create using parts and constraints
- show the model
- network view
- basic parts view. Matt suggested being able to view both strands and flip them around.
- take in conditions for synthetic evolution and send models to the evolutionary algorithm
- output the sequence in a standard format
User Stories
Parts Repository
- The program must display a list of all available parts. The program requests a hierarchical list of all part names, part types, and their part ids from the Parts Repository (PR). Upon receiving the list, the program sorts the parts into different categories based on the part type, chooses a display icon for each type, and presents the list of part names with their part type icons to the user.
- The user wishes to change the type of chassis. The program retrieves a list of all chassis names and chassis ids from the PR and presents it to the user.
- The program must display a list of all types of parts. It requests a list of all part types and their part type ids from the PR. Then it displays the list of part names to the user.
- The program must display all of the information about a single part. It gives the part id to the PR and receives a full description of the part in return. It then displays the part information to the user.
- The program must save a CellML model of a part. It gives the part id to the PR and receives a segment of CellML in return. It then saves the CellML to file.
Constraints Repository
- The program must show all possible interactions to a particular part. It passes the part id to the Constraints Repository and receives a list of all possible interaction ids in return. It then retrieves the details of each interaction with the interaction id. It then shows the possible interactions to the user.
Evolutionary Algorithm
- The user finally wants to evolve her model in silico. The program collects the starting model, composed of a series of part ids, and the proposed behaviour in the form of a truth table, and gives it to the Evolutionary Algorithm (EA) for evolution. The program shows a “busy working” screen while the Evolutionary Algorithm computes the best model. When the EA finishes, the program receives the proposed best model(s) and displays them, along with their behaviour, to the user.
Outcome
Darwin in Silico (DiS) is the Workbench: the graphical user interface (GUI) for the design of the genetic construct that was engineered into B. subtilis. The evolutionary algorithm, parts repository and the constraints repository work behind the scenes of this interface in the design of the construct.
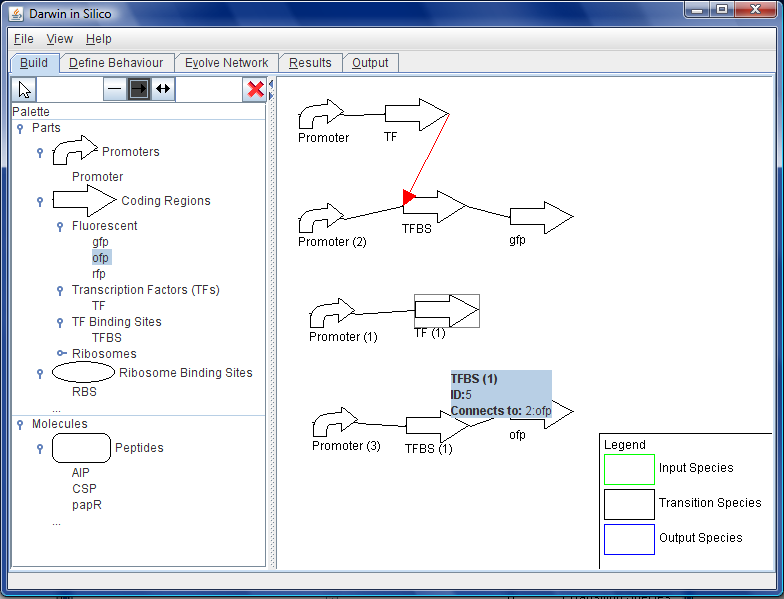
The user can design a starting point for the genetic construct in the centre window of the GUI by dragging and dropping parts into place. All of the two-component genes are available for this process, as are a large number of promoters, ribosome binding sites, transcription factors and sigma factors. These parts are accessed from the parts repository database and built into a preliminary construct.
The constraints repository aids in the building of this construct by not allowing parts that are not compatible to be dropped in place next to each other. Where parts are compatible with each other, constraint models are incorporated into the construct that define the interactions between these.
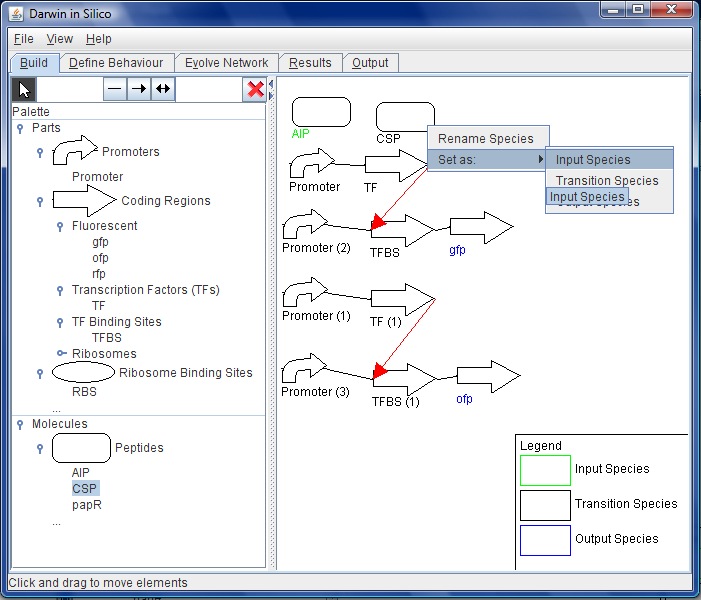
The user can also map system inputs to system outputs that can be used by the EA. For example, the user could specify that in the presence of a certain set of quorum sensing peptides that a certain fluorescent protein would light up. The user can then run the EA through DiS, leading to the mutation of the hidden layer and the output of the hidden layer model that fulfils the output requirements the user has set.
All of the different software components were supposed to communicate through DiS. Each team member was responsible for the construction of their component. The outcome of this was a working build, where the evolutionary algorithm could communicate directly with the parts repository, but not with DiS. The components were simply not ready to communicate with each other. Once some more work has been carried out on the various components of the overall project, DiS will be functional.
Contributors
Lead: Morgan Taschuk
 "
"