Team:Calgary Software/Project
From 2008.igem.org
(→Graphical User Interface) |
(→Graphical User Interface) |
||
| Line 104: | Line 104: | ||
[[Image:GUIPic2.jpg|thumb|More advanced users can re-configure the initial settings]] | [[Image:GUIPic2.jpg|thumb|More advanced users can re-configure the initial settings]] | ||
[[Image:GUIPic3.jpg|thumb|More advanced users can re-configure the initial settings]] | [[Image:GUIPic3.jpg|thumb|More advanced users can re-configure the initial settings]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
Users that are more familiar with the system or with the principles of Evolutionary Strategies can edit the initial settings according to the specific needs of the problem. This gives much more flexibility in terms of search methods and allows more advanced users to take advantage of their knowledge for this specific problem. | Users that are more familiar with the system or with the principles of Evolutionary Strategies can edit the initial settings according to the specific needs of the problem. This gives much more flexibility in terms of search methods and allows more advanced users to take advantage of their knowledge for this specific problem. | ||
| Line 109: | Line 117: | ||
[[Image:GUIPic4.jpg|thumb|Users can aid the system's search algorithm by giving it some previous knowledge they hold]] | [[Image:GUIPic4.jpg|thumb|Users can aid the system's search algorithm by giving it some previous knowledge they hold]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
The interface goes even further in allowing users to harness previous knowledge for any specific bio-circuit they wish to design. Users may specify an "embryo" circuit for the system to start its optimization from. This circuit will be used as the base template for the evolution an could save some time and effort for the system, and also for the user. | The interface goes even further in allowing users to harness previous knowledge for any specific bio-circuit they wish to design. Users may specify an "embryo" circuit for the system to start its optimization from. This circuit will be used as the base template for the evolution an could save some time and effort for the system, and also for the user. | ||
| Line 114: | Line 141: | ||
[[Image:GUIPic5.jpg|thumb|The parts are chosen from a graphical list that represents the registry]] | [[Image:GUIPic5.jpg|thumb|The parts are chosen from a graphical list that represents the registry]] | ||
[[Image:GUIPic6.jpg|thumb|Each part's characteristics can be viewed on demand]] | [[Image:GUIPic6.jpg|thumb|Each part's characteristics can be viewed on demand]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
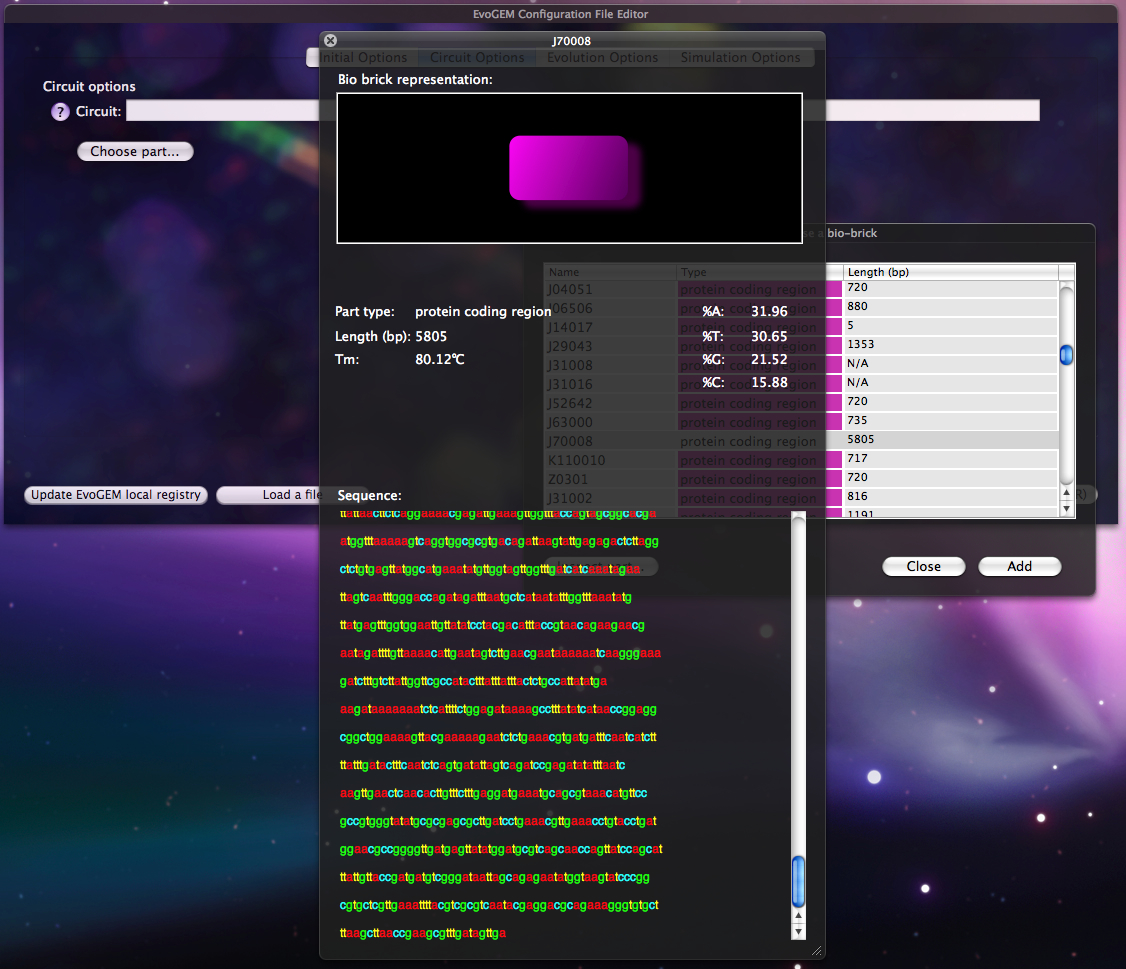
Users can choose the parts they wish to include in the "embryo" circuit from a list that reflects the results obtained from the registry query by Perl. For every part, there is also the option to inspect its characteristics such as: sequence, different base constituency , melting temperature and more. | Users can choose the parts they wish to include in the "embryo" circuit from a list that reflects the results obtained from the registry query by Perl. For every part, there is also the option to inspect its characteristics such as: sequence, different base constituency , melting temperature and more. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
</div> | </div> | ||
Revision as of 23:35, 29 October 2008

|
| Home | The Team | The Project | Modeling | Notebook |
|---|
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
Contents |
Introduction
We created EvoGEM, a software program that uses the registry of genetic parts provided by the iGEM competition. In this design, we used evolutionary and genetic strategies, which are useful modeling methods, especially when coupled with agent-based designs.
Paradigm of Evolution
EvoGEM uses the paradigm of evolution to select efficient designs and produce a product or output that is generated independently by the system. EvoGEM uses the strategies of genetics and evolution to simulate an environment inside of a prokaryotic cell. This entails various events and structures that are present inside the organism, such as:
- RNA polymerase
- messenger RNA
- ribosomes
- transcription and translation.
Goals and Achievements
The last team from University of Calgary presented EvoGEM during the 2007 iGEM Jamboree, where it sparked a lot of interest. This summer, our team has expanded EvoGEM by:
- improving EvoGEM's fitness function.
- introducing more complex pattern recognition.
- testing the system under a much larger search space.
Our goal was to create a system sophisticated enough to rebuild working designs from previous years' teams' projects, as well as intelligent enough to simulate successes and failures of functional and dysfunctional systems, respectively. We achieved our goals by:
- building Perl scripts that support EvoGEM's need for a flat file registry.
- creating a graphical user interface (GUI) to make the software-user interaction easier.
- creating a simulation of the processes in the cell such as transcription and translation.
Essentially, this software will allow users to determine whether a specific circuit is feasible before they experimentally test it. This will reduce financial and time constraints associated with traditional lab work. The user will only need to invest time and money into those circuits that our program selects as functional.
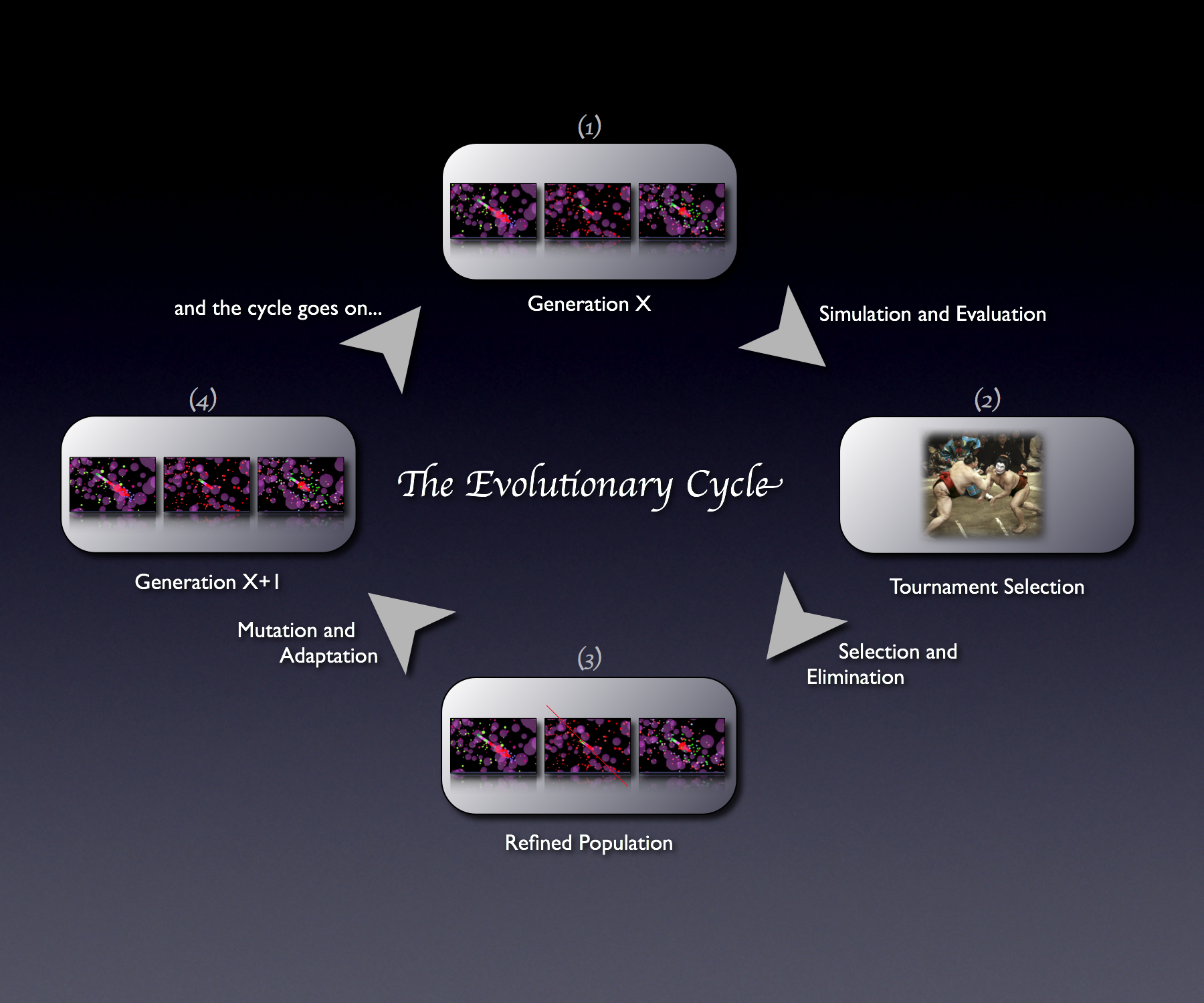
Evolutionary Algorithm
Evolution involves the changes of inherited traits in a population for successive generations. Each generation carries genetic information, expressing certain characteristics. Mutation enables manipulation of these traits as well as genetic recombination. Evolution is a result of heritable traits becoming more prevalent or rare.
Agent-based modeling is a computational method that replicates the behavior and interaction of individual components of a network such that their overall effect on the system can be observed. This involves many different aspects, including:
- game theory
- evolutionary programming
- complex systems
- emergence
Data Retrieval and Storage
- its type.
- its function.
- whether it codes for a protein.
- how well the part works.
If the part coded for a protein, we retrieved its DNA sequence. We, then, found its amino acid sequence (using the BLAST algorithm) from UniProt, which is a large database of proteins. If the protein catalyzed an associated prosthetic or biochemical reaction, we retrieved additional information from ChemSpider - a chemical database - to find the data characterizing any compounds that the particular enzymatic protein catalyzes. Finally, we stored the data in a database during run-time of EvoGEM.
Modeling
Graphical User Interface
Programs are only useful if the user is able to understand and use it. Working through a terminal or command prompt may be easy and simple for some, but for the majority of people, this is not something people can easily use. As a result, a Graphical User Interface (GUI) was created to allow anyone to use EvoGEM. Since the intent of EvoGEM was to help anyone dealing with synthetic biology, this GUI is essential.
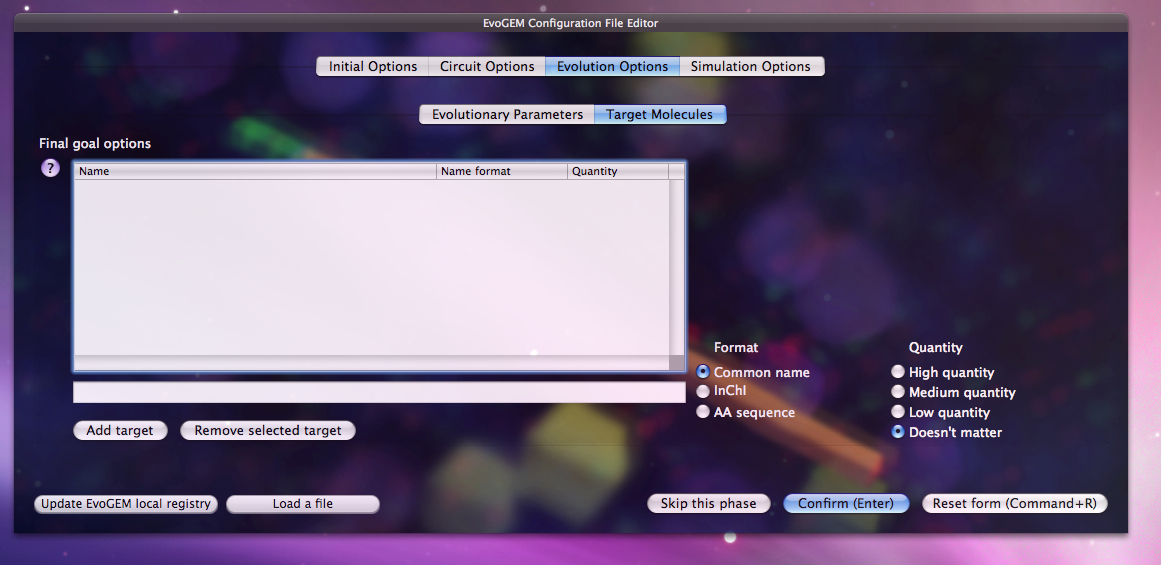
When first faced with the GUI, the user is asked to enter his or her desired products and their relative quantities. That is, in essence the only input the system needs. Everything else is pre-configured for the system and the user can run the software to produce the circuits that fulfill his or her needs.
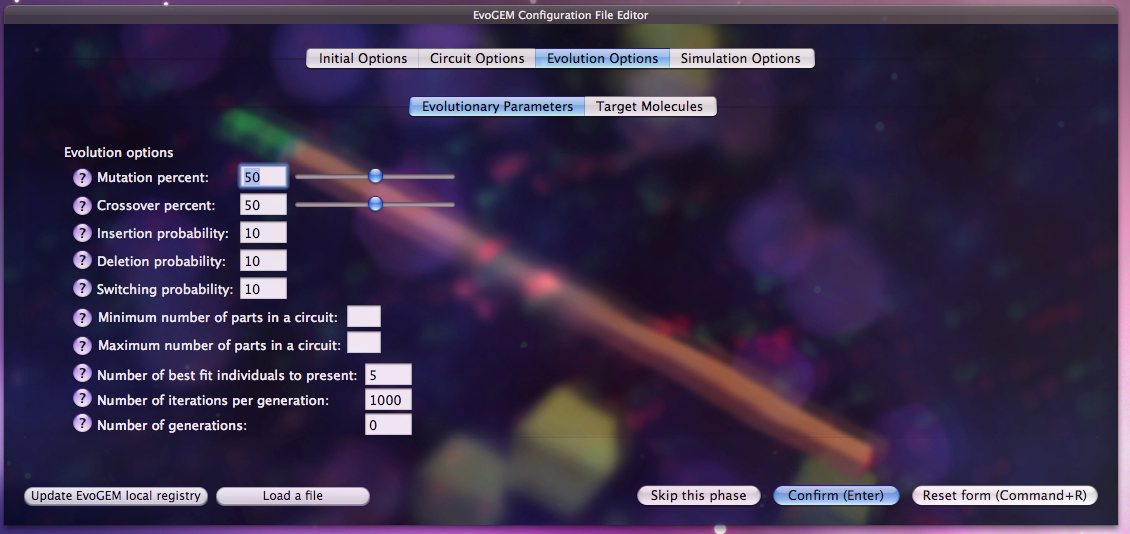
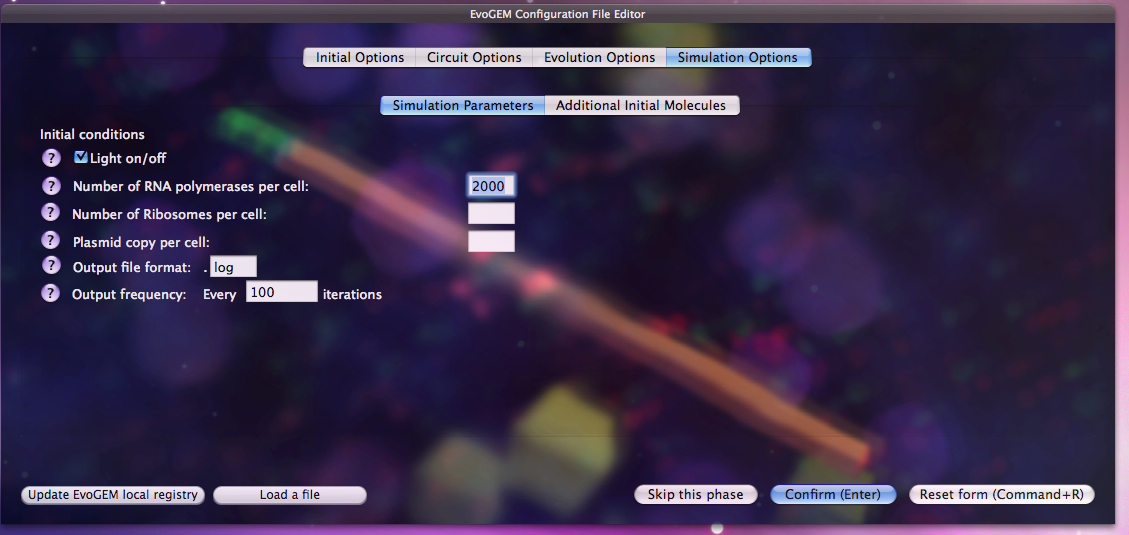
Users that are more familiar with the system or with the principles of Evolutionary Strategies can edit the initial settings according to the specific needs of the problem. This gives much more flexibility in terms of search methods and allows more advanced users to take advantage of their knowledge for this specific problem.
The interface goes even further in allowing users to harness previous knowledge for any specific bio-circuit they wish to design. Users may specify an "embryo" circuit for the system to start its optimization from. This circuit will be used as the base template for the evolution an could save some time and effort for the system, and also for the user.
Users can choose the parts they wish to include in the "embryo" circuit from a list that reflects the results obtained from the registry query by Perl. For every part, there is also the option to inspect its characteristics such as: sequence, different base constituency , melting temperature and more.
| Evolutionary Algorithm | Data Retrieval | Modeling | Graphical User Interface |
|---|
| Home | The Team | The Project | Modeling | Notebook |
|---|
 "
"